1LAM
 
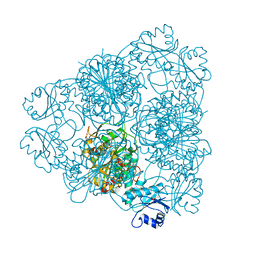 | | LEUCINE AMINOPEPTIDASE (UNLIGATED) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, CARBONATE ION, LEUCINE AMINOPEPTIDASE, ... | | 著者 | Straeter, N, Lipscomb, W.N. | | 登録日 | 1995-08-11 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1LCP
 
 | |
4I07
 
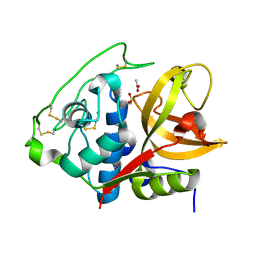 | | Structure of mature form of cathepsin B1 from Schistosoma mansoni | | 分子名称: | ACETATE ION, CHLORIDE ION, Cathepsin B-like peptidase (C01 family) | | 著者 | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | 登録日 | 2012-11-16 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Activation route of the Schistosoma mansoni cathepsin B1 drug target: structural map with a glycosaminoglycan switch
Structure, 22, 2014
|
|
1L9M
 
 | |
1M9U
 
 | |
2EST
 
 | |
2Z71
 
 | | Structure of truncated mutant CYS1GLY of penicillin V acylase from bacillus sphaericus co-crystallized with penicillin V | | 分子名称: | (2S,5R,6R)-3,3-DIMETHYL-7-OXO-6-(2-PHENOXYACETAMIDO)-4-THIA-1- AZABICYCLO(3.2.0)HEPTANE-2-CARBOXYLIC ACID, Penicillin acylase | | 著者 | Pathak, M.C, Brannigan, J, Dodson, G.G, Suresh, C.G. | | 登録日 | 2007-08-10 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Studies on the catalysis and post translational processing of penicillin V acylase
To be published
|
|
3LQM
 
 | | Structure of the IL-10R2 Common Chain | | 分子名称: | GLYCEROL, Interleukin-10 receptor subunit beta, SULFATE ION | | 著者 | Yoon, S.I, Walter, M.R. | | 登録日 | 2010-02-09 | | 公開日 | 2010-05-26 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structure and mechanism of receptor sharing by the IL-10R2 common chain.
Structure, 18, 2010
|
|
5T87
 
 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | 分子名称: | CdiA toxin, CdiI immunity protein | | 著者 | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-09-06 | | 公開日 | 2017-09-13 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
2WUB
 
 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp | | 分子名称: | FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, FAB FRAGMENT FAB40.DELTATRP LIGHT CHAIN, HEPATOCYTE GROWTH FACTOR ACTIVATOR LONG CHAIN, ... | | 著者 | Ganesan, R, Eigenbrot, C, Shia, S. | | 登録日 | 2009-10-01 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
7LPR
 
 | |
1PBI
 
 | |
1YPP
 
 | | ACID ANHYDRIDE HYDROLASE | | 分子名称: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | 著者 | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | 登録日 | 1996-05-29 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
6LPR
 
 | |
4TRA
 
 | |
3HER
 
 | | Human prion protein variant F198S with V129 | | 分子名称: | CADMIUM ION, Major prion protein | | 著者 | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | 登録日 | 2009-05-10 | | 公開日 | 2010-01-12 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HJX
 
 | | Human prion protein variant D178N with V129 | | 分子名称: | CADMIUM ION, CHLORIDE ION, Major prion protein | | 著者 | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | 登録日 | 2009-05-22 | | 公開日 | 2010-01-12 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HJ5
 
 | | Human prion protein variant V129 domain swapped dimer | | 分子名称: | Major prion protein | | 著者 | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | 登録日 | 2009-05-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
220L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
200L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
223L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
222L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
