7VUU
 
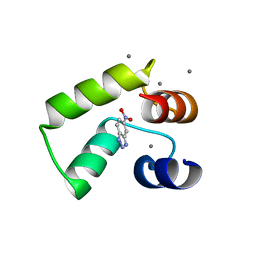 | | Crystal structure of AlleyCat10 with inhibitor | | 分子名称: | 5-nitro-1H-benzotriazole, AlleyCat, CALCIUM ION | | 著者 | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | 登録日 | 2021-11-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7WCG
 
 | |
7VUT
 
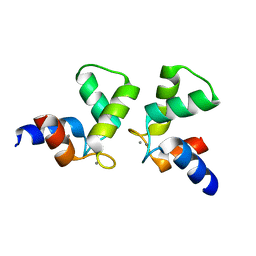 | | Crystal structure of AlleyCat10 | | 分子名称: | AlleyCat10, CALCIUM ION | | 著者 | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | 登録日 | 2021-11-04 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | NMR-guided directed evolution.
Nature, 610, 2022
|
|
5WAH
 
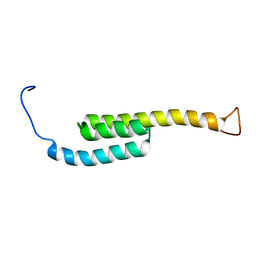 | | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN | | 分子名称: | IgA FC receptor | | 著者 | ELETSKY, A, CHEN, C, FONG, J.J, NIZET, V, VARKI, A, PRESTEGARD, J.H. | | 登録日 | 2017-06-26 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN
To Be Published
|
|
6KCZ
 
 | |
6MPP
 
 | |
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | 分子名称: | RIBONUCLEASE | | 著者 | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | 登録日 | 1998-05-05 | | 公開日 | 1998-10-14 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
7A4L
 
 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | 分子名称: | IRON/SULFUR CLUSTER, PioC | | 著者 | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | 登録日 | 2020-08-19 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
6QYW
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - nisin ring A | | 分子名称: | ILE-DBU-DAL-ILE-DHA-LEU-CYS-ALA | | 著者 | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | 登録日 | 2019-03-09 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-10-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYV
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A (Ser2, Ala5, Ala8) analogue | | 分子名称: | PHE-SER-DAL-LEU-ALA-LEU-CYS-ALA | | 著者 | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | 登録日 | 2019-03-09 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-10-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
1AYG
 
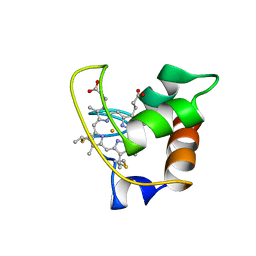 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | 分子名称: | CYTOCHROME C-552, HEME C | | 著者 | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | 登録日 | 1997-11-04 | | 公開日 | 1998-11-25 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
7P51
 
 | |
7N45
 
 | |
6QYU
 
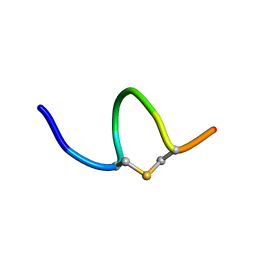 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A | | 分子名称: | PHE-DHA-DAL-LEU-DHA-LEU-CYS-ALA | | 著者 | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | 登録日 | 2019-03-09 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | 分子名称: | Beta-endorphin | | 著者 | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | 登録日 | 2020-01-05 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6QYT
 
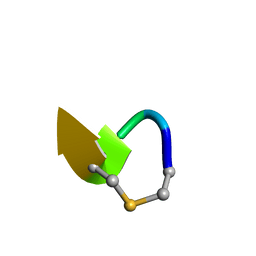 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A truncated analogue | | 分子名称: | DAL-LEU-SER-LEU-CYS-ALA | | 著者 | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | 登録日 | 2019-03-09 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
5LCH
 
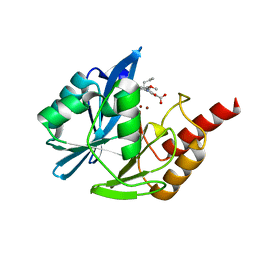 | | VIM-2 metallo-beta-lactamase in complex with (S)-1-allyl-2-(3-methoxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 42) | | 分子名称: | (1~{S})-2-(3-methoxyphenyl)-3-oxidanylidene-1-prop-2-enyl-1~{H}-isoindole-4-carboxylic acid, Metallo-beta-lactamase VIM-2, ZINC ION | | 著者 | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2016-06-21 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LCA
 
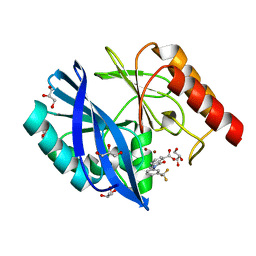 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-(3-(trifluoromethyl)phenyl)isoindoline-4-carboxylic acid (compound 17) | | 分子名称: | 3-oxidanylidene-2-[3-(trifluoromethyl)phenyl]-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase VIM-2, ... | | 著者 | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2016-06-20 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5VX7
 
 | |
5LCF
 
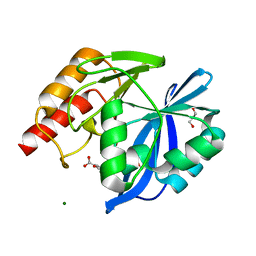 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-phenylisoindoline-4-carboxylic acid (compound 30) | | 分子名称: | 3-oxidanylidene-2-phenyl-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2016-06-21 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LE1
 
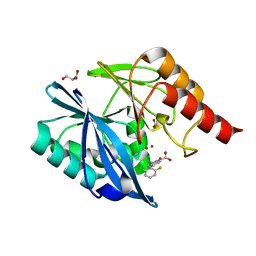 | | VIM-2 metallo-beta-lactamase in complex with 2-(2-chloro-6-fluorobenzyl)-3-oxoisoindoline-4-carboxylic acid (compound 16) | | 分子名称: | 2-[(2-chloranyl-6-fluoranyl-phenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, GLYCEROL, ... | | 著者 | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2016-06-29 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
5LM6
 
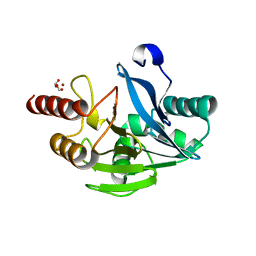 | | VIM-2 metallo-beta-lactamase in complex with 2-(3-fluoro-4-hydroxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 35) | | 分子名称: | 2-(3-fluoranyl-4-oxidanyl-phenyl)-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, Metallo-beta-lactamase VIM-2, ... | | 著者 | Li, G.-B, Brem, J, Someya, H, McDonough, M.A, Schofield, C.J. | | 登録日 | 2016-07-29 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
1A91
 
 | | SUBUNIT C OF THE F1FO ATP SYNTHASE OF ESCHERICHIA COLI; NMR, 10 STRUCTURES | | 分子名称: | F1FO ATPASE SUBUNIT C | | 著者 | Girvin, M.E, Rastogi, V.K, Abildgaard, F, Markley, J.L, Fillingame, R.H. | | 登録日 | 1998-04-15 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase.
Biochemistry, 37, 1998
|
|
1AH2
 
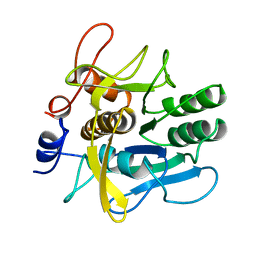 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | 分子名称: | SERINE PROTEASE PB92 | | 著者 | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | 登録日 | 1997-04-11 | | 公開日 | 1998-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
1B22
 
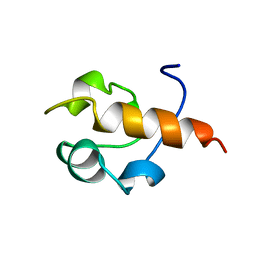 | | RAD51 (N-TERMINAL DOMAIN) | | 分子名称: | DNA REPAIR PROTEIN RAD51 | | 著者 | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
