3MIG
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite, obtained in the presence of substrate | | 分子名称: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | 著者 | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | 登録日 | 2010-04-10 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
5BW9
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | 著者 | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | 登録日 | 2015-06-06 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (7 Å) | | 主引用文献 | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
5WQD
 
 | |
5D80
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | 著者 | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | 登録日 | 2015-08-14 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (6.202 Å) | | 主引用文献 | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
8VSU
 
 | | Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, Isoform 3 of STE20-related kinase adapter protein alpha, ... | | 著者 | Chan, L.M, Courteau, B.J, Verba, K.A. | | 登録日 | 2024-01-24 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | High-resolution single-particle imaging at 100-200 keV with the Gatan Alpine direct electron detector.
J.Struct.Biol., 216, 2024
|
|
7ZQY
 
 | |
7ZR1
 
 | | Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure) | | 分子名称: | DH domain-containing protein, Double-strand break repair protein, FHA domain-containing protein, ... | | 著者 | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | 登録日 | 2022-05-03 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
2DKH
 
 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, in complex with the substrate | | 分子名称: | 3-HYDROXYBENZOIC ACID, 3-hydroxybenzoate hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | 登録日 | 2006-04-11 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
8CF2
 
 | | Solution structure of the RNA helix formed by the 5'-end of U1 snRNA and an A-1 bulged 5'-splice site in complex with SMN-CY | | 分子名称: | 4-[(3~{S})-3-ethylpiperazin-1-yl]-2-fluoranyl-~{N}-(2-methylimidazo[1,2-a]pyrazin-6-yl)benzamide, RNA (5'-R(P*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | 著者 | Malard, F, Marquevielle, J, Campagne, S. | | 登録日 | 2023-02-02 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
3WGW
 
 | | Structure of PCNA bound to a small molecule inhibitor | | 分子名称: | 4-{4-[(2S)-2-amino-3-hydroxypropyl]-2,6-diiodophenoxy}phenol, Proliferating cell nuclear antigen, SULFATE ION | | 著者 | Hashimoto, H. | | 登録日 | 2013-08-12 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A small molecule inhibitor of monoubiquitinated Proliferating Cell Nuclear Antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin.
J.Biol.Chem., 289, 2014
|
|
6ETJ
 
 | | HUMAN PFKFB3 IN COMPLEX WITH KAN0438241 | | 分子名称: | 4-[[3-(5-fluoranyl-2-oxidanyl-phenyl)phenyl]sulfonylamino]-2-oxidanyl-benzoic acid, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | 著者 | Gustafsson, N.M.S, Lundback, T, Farnegardh, K, Groth, P, Wiitta, E, Jonsson, M, Hallberg, K, Pennisi, R, Huguet Ninou, A, Martinsson, J, Norstrom, C, Schultz, J, Andersson, M, Markova, N, Marttila, P, Norin, M, Olin, T, Helleday, T. | | 登録日 | 2017-10-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun, 9, 2018
|
|
3DSC
 
 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | 分子名称: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | 著者 | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | 登録日 | 2008-07-11 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3E8K
 
 | |
3DSD
 
 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | 分子名称: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | 著者 | Williams, R.S, Moiani, D, Tainer, J.A. | | 登録日 | 2008-07-11 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3DAB
 
 | |
1HJ4
 
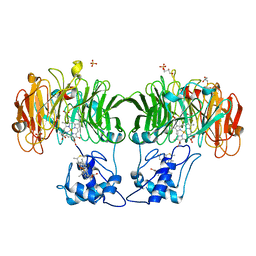 | |
1HJ3
 
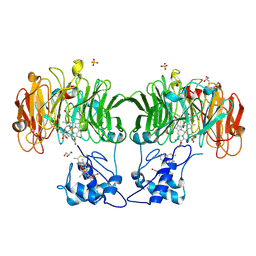 | |
1HJ5
 
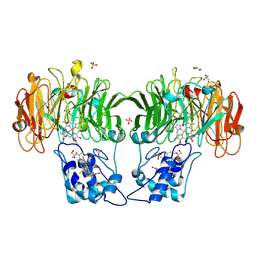 | |
1LB7
 
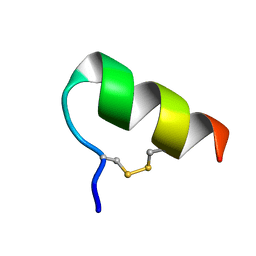 | | IGF-F1-1, A PEPTIDE ANTAGONIST OF IGF-1 | | 分子名称: | IGF-1 ANTAGONIST F1-1 | | 著者 | Deshayes, K, Schaffer, M.L, Skelton, N.J, Nakamura, G.R, Kadkhodayan, S, Sidhu, S.S. | | 登録日 | 2002-04-02 | | 公開日 | 2002-06-19 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rapid identification of small binding motifs with high-throughput phage display: discovery of peptidic antagonists of IGF-1 function.
Chem.Biol., 9, 2002
|
|
1MK6
 
 | | SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT MISPAIRED WITH DEOXYADENOSINE | | 分子名称: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*AP*GP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | 著者 | Giri, I, Johnston, D.S, Stone, M.P. | | 登録日 | 2002-08-28 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | MISPAIRING OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT WITH DEOXYADENOSINE RESULTS IN EXTRUSION OF THE MISMATCHED DA TOWARD THE MAJOR GROOVE
Biochemistry, 41, 2002
|
|
7FDA
 
 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | 分子名称: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | 分子名称: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDB
 
 | | CryoEM Structures of Reconstituted V-ATPase,State2 | | 分子名称: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDE
 
 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | 分子名称: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | 登録日 | 2021-07-16 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
5YUI
 
 | | CO2 release in human carbonic anhydrase II crystals: reveal histidine 64 and solvent dynamics | | 分子名称: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | 著者 | Kim, C.U, Park, S.Y, McKenna, R. | | 登録日 | 2017-11-22 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
