7KDK
 
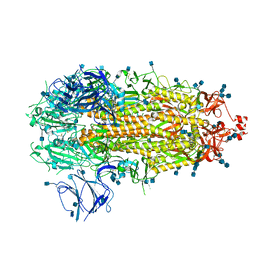 | |
7K9Z
 
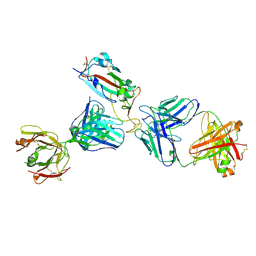 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | 著者 | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
7KE6
 
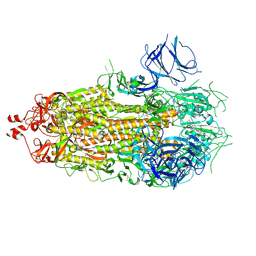 | |
7KEC
 
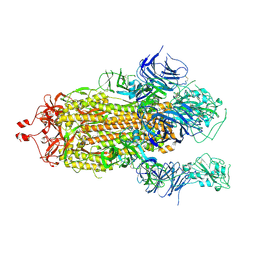 | |
7KE7
 
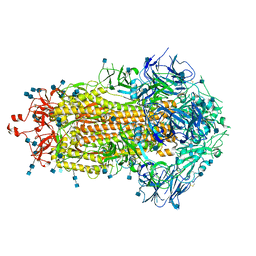 | |
7LFV
 
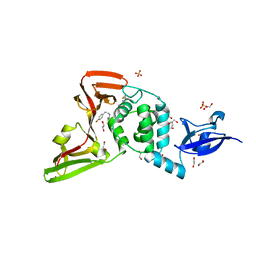 | |
7LFU
 
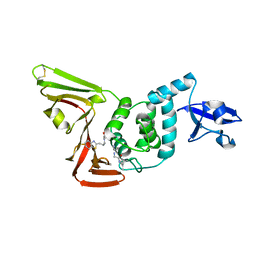 | |
7LTN
 
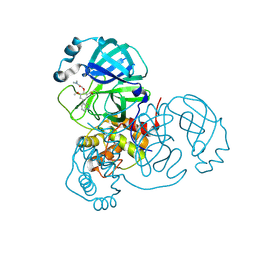 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | 分子名称: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | 著者 | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | 登録日 | 2021-02-19 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LLZ
 
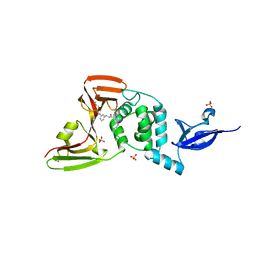 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-69 | | 分子名称: | N-[(1R)-1-(3-{5-[(acetylamino)methyl]thiophen-2-yl}phenyl)ethyl]-5-[(azetidin-3-yl)amino]-2-methylbenzamide, Non-structural protein 3, SULFATE ION, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-02-04 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LBR
 
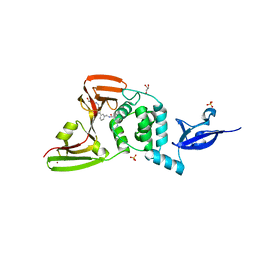 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-89 | | 分子名称: | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1S,3R)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide, GLYCEROL, Non-structural protein 3, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-01-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LBS
 
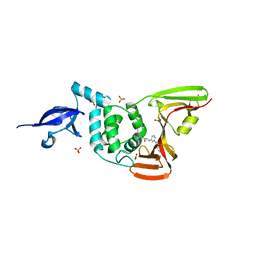 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-24 | | 分子名称: | 5-[(azetidin-3-yl)amino]-2-methyl-N-[(1R)-1-(3-{5-[(pyrrolidin-1-yl)methyl]thiophen-2-yl}phenyl)ethyl]benzamide, BORIC ACID, GLYCEROL, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-01-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LS9
 
 | |
7LXW
 
 | |
7LY0
 
 | |
7LXZ
 
 | |
7LY2
 
 | |
7LXX
 
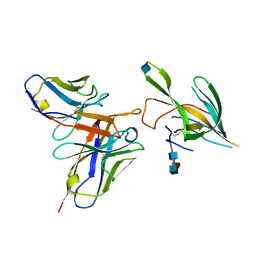 | |
7LXY
 
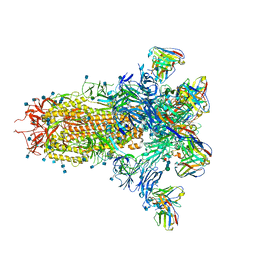 | |
7LY3
 
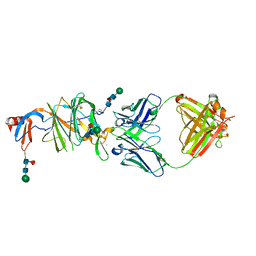 | |
7LMC
 
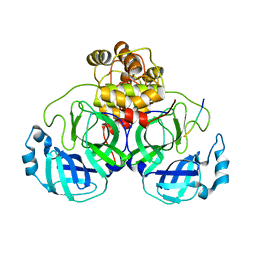 | |
7K0E
 
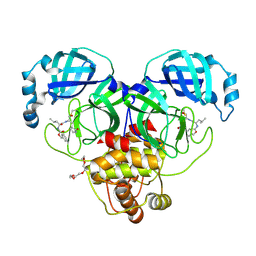 | | 1.90 A resolution structure of SARS-CoV-2 3CL protease in complex with deuterated GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LHQ
 
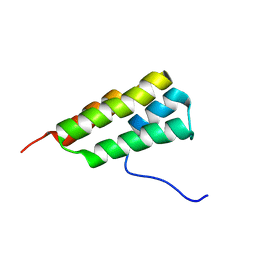 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | 分子名称: | Non-structural protein 7 | | 著者 | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | 登録日 | 2021-01-26 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
7M1Y
 
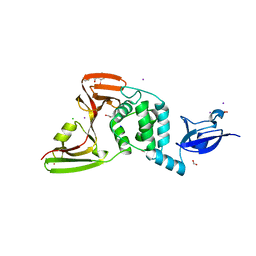 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Maltseva, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-03-15 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with ebselen
to be published
|
|
7M4R
 
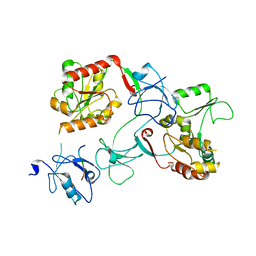 | |
7KE9
 
 | |
