6VLS
 
 | | Structure of C-terminal fragment of Vip3A toxin | | 分子名称: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Vip3Aa | | 著者 | Jiang, K, Zhang, Y, Chen, Z, Gao, X. | | 登録日 | 2020-01-25 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and Functional Insights into the C-terminal Fragment of Insecticidal Vip3A Toxin ofBacillus thuringiensis.
Toxins, 12, 2020
|
|
5C71
 
 | | The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide | | 分子名称: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Glucuronidase, alpha-D-glucopyranuronic acid | | 著者 | Sun, H.L, Lv, B, Huang, S, Li, C, Jiang, T. | | 登録日 | 2015-06-24 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structure-guided engineering of the substrate specificity of a fungal beta-glucuronidase toward triterpenoid saponins.
J.Biol.Chem., 293, 2018
|
|
3B4Y
 
 | | FGD1 (Rv0407) from Mycobacterium tuberculosis | | 分子名称: | CITRATE ANION, COENZYME F420, PROBABLE F420-DEPENDENT GLUCOSE-6-PHOSPHATE DEHYDROGENASE FGD1 | | 著者 | Bashiri, G, Squire, C.J, Moreland, N.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2007-10-25 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding
J.Biol.Chem., 283, 2008
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HJ9
 
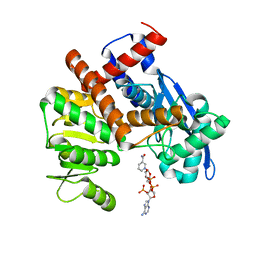 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HOW
 
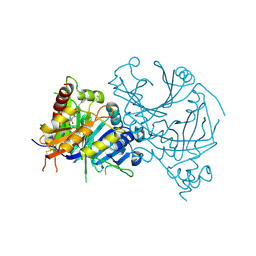 | | Crystal structure of AtHPPD-Y191052 complex | | 分子名称: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(2-phenylethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, G.-F, Lin, H.-Y, Dong, J. | | 登録日 | 2022-12-11 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-02-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
8HJ3
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
6CVY
 
 | |
8HHO
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HIQ
 
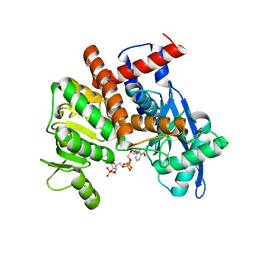 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-21 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
4WZJ
 
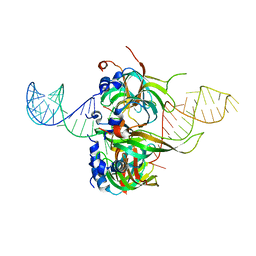 | | Spliceosomal U4 snRNP core domain | | 分子名称: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | 著者 | Leung, A.K.W, Nagai, K, Li, J. | | 登録日 | 2014-11-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
8HIZ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
6W70
 
 | | Crystal Structure of apixaban-bound ABLE | | 分子名称: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, ACETATE ION, De novo designed ABLE, ... | | 著者 | Polizzi, N.F. | | 登録日 | 2020-03-18 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.296 Å) | | 主引用文献 | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
4X6A
 
 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Wang, L, Chong, J, Wang, D. | | 登録日 | 2014-12-07 | | 公開日 | 2015-02-04 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (3.96 Å) | | 主引用文献 | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1G8Q
 
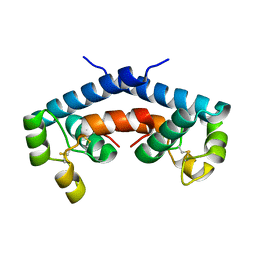 | | CRYSTAL STRUCTURE OF HUMAN CD81 EXTRACELLULAR DOMAIN, A RECEPTOR FOR HEPATITIS C VIRUS | | 分子名称: | CD81 ANTIGEN, EXTRACELLULAR DOMAIN | | 著者 | Kitadokoro, K, Bolognesi, M, Bordo, D, Grandi, G, Galli, G, Petracca, R, Falugi, F. | | 登録日 | 2000-11-20 | | 公開日 | 2001-02-21 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs.
EMBO J., 20, 2001
|
|
5CGD
 
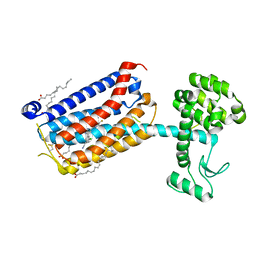 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile - (HTL14242) | | 分子名称: | 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, OLEIC ACID | | 著者 | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | 登録日 | 2015-07-09 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
5CGC
 
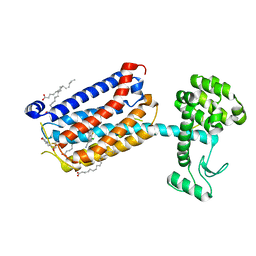 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | 著者 | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | 登録日 | 2015-07-09 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
2EQ5
 
 | |
5CHO
 
 | |
8H97
 
 | | GH86 agarase Aga86A_Wa | | 分子名称: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | 著者 | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | 登録日 | 2022-10-25 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
6CCN
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-2,4-dihydroxy-N-(2-(4-hydroxy-1H-benzo[d]imidazol-2-yl)ethyl)-3,3-dimethylbutanamide | | 分子名称: | (2R)-2,4-dihydroxy-N-[2-(7-hydroxy-1H-benzimidazol-2-yl)ethyl]-3,3-dimethylbutanamide, Phosphopantetheine adenylyltransferase, SULFATE ION | | 著者 | Mamo, M, Appleton, B.A. | | 登録日 | 2018-02-07 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
5CLD
 
 | |
5CLA
 
 | |
1YQK
 
 | | Human 8-oxoguanine glycosylase crosslinked with guanine containing DNA | | 分子名称: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*G)-3', 5'-D(P*CP*AP*GP*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | 著者 | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | 登録日 | 2005-02-01 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
