3QF7
 
 | |
3QG5
 
 | |
7Q2Z
 
 | |
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | 登録日 | 2019-12-19 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.58 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRF
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N. | | 登録日 | 2019-12-18 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.106 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N. | | 登録日 | 2019-12-19 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (5.74 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | 分子名称: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | 著者 | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | 登録日 | 2019-12-20 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
7U1A
 
 | | RFC:PCNA bound to dsDNA with a ssDNA gap of six nucleotides | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | 著者 | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | 登録日 | 2022-02-20 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7U19
 
 | | RFC:PCNA bound to nicked DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA, MAGNESIUM ION, ... | | 著者 | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | 登録日 | 2022-02-20 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7U1P
 
 | | RFC:PCNA bound to DNA with a ssDNA gap of five nucleotides | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | 著者 | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | 登録日 | 2022-02-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
8ITE
 
 | |
1EM8
 
 | | Crystal structure of chi and psi subunit heterodimer from DNA POL III | | 分子名称: | DNA POLYMERASE III CHI SUBUNIT, DNA POLYMERASE III PSI SUBUNIT | | 著者 | Gulbis, J.M, Finkelstein, J, O'Donnell, M, Kuriyan, J. | | 登録日 | 2000-03-16 | | 公開日 | 2003-08-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex.
Eur.J.Biochem., 271, 2004
|
|
8DR0
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | 分子名称: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQZ
 
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | 分子名称: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQX
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR6
 
 | | Closed state of RFC:PCNA bound to a nicked dsDNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (32-MER), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
5OHQ
 
 | |
5OIK
 
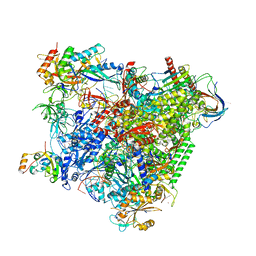 | | Structure of an RNA polymerase II-DSIF transcription elongation complex | | 分子名称: | DNA (43-MER), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | 著者 | Bernecky, C, Plitzko, J.M, Cramer, P. | | 登録日 | 2017-07-18 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OHO
 
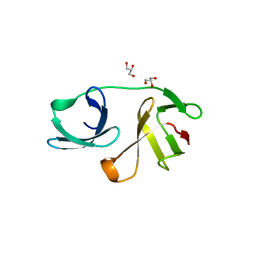 | |
6VVO
 
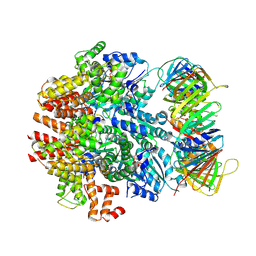 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | 登録日 | 2020-02-18 | | 公開日 | 2020-02-26 | | 最終更新日 | 2020-03-25 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
8GJ3
 
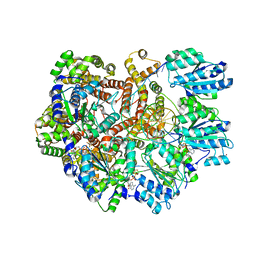 | |
6KUF
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with glucose. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-09-02 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6L1J
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4'-nitrophenyl thiolaminaritrioside | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-09-29 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LC5
 
 | | Crystal structure of barley exohydrolaseI W434F in complex with 4'-nitrophenyl thiolaminaribioside | | 分子名称: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-11-17 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LBB
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-11-14 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
