7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | 分子名称: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | 著者 | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | 登録日 | 2021-04-13 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
8R8N
 
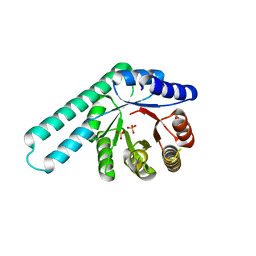 | |
8R8O
 
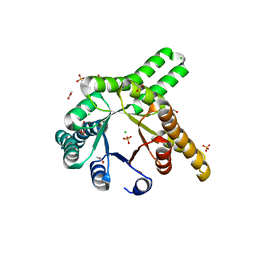 | | Hallucinated de novo TIM barrel with three helical extensions - HalluTIM3-1 | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Beck, J, Shanmugaratnam, S, Hocker, B. | | 登録日 | 2023-11-29 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Diversifying de novo TIM barrels by hallucination.
Protein Sci., 33, 2024
|
|
3P6J
 
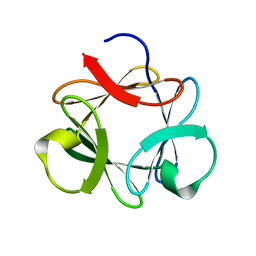 | |
2MTQ
 
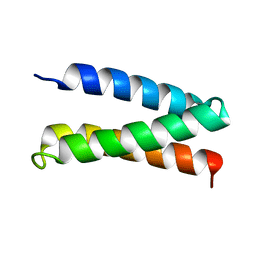 | | Solution Structure of a De Novo Designed Peptide that Sequesters Toxic Heavy Metals | | 分子名称: | Designed Peptide | | 著者 | Plegaria, J.S, Zuiderweg, E.R, Stemmler, T.L, Pecoraro, V.L. | | 登録日 | 2014-08-28 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Apoprotein Structure and Metal Binding Characterization of a de Novo Designed Peptide, alpha 3DIV, that Sequesters Toxic Heavy Metals.
Biochemistry, 54, 2015
|
|
6W40
 
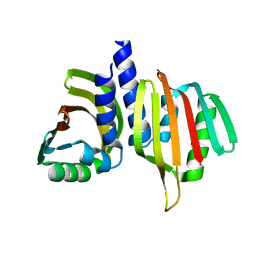 | |
6LLQ
 
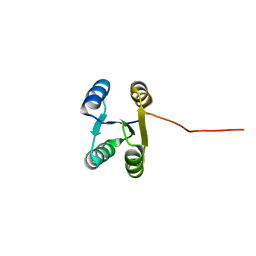 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | 分子名称: | VAL88 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | 登録日 | 2019-12-23 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | 登録日 | 2016-10-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-10-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TRV
 
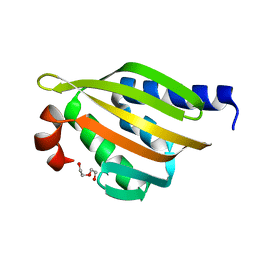 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-10-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | 分子名称: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | 登録日 | 2020-05-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | 分子名称: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | 登録日 | 2020-05-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
3VJF
 
 | | Crystal structure of de novo 4-helix bundle protein WA20 | | 分子名称: | POTASSIUM ION, WA20 | | 著者 | Arai, R, Kimura, A, Kobayashi, N, Matsuo, K, Sato, T, Wang, A.F, Platt, J.M, Bradley, L.H, Hecht, M.H. | | 登録日 | 2011-10-18 | | 公開日 | 2012-03-28 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20
J.Phys.Chem.B, 116, 2012
|
|
7BEY
 
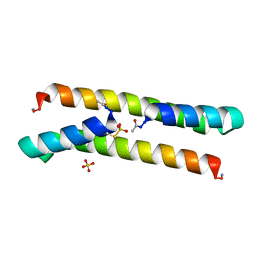 | | Het-N2-SO3- - De novo designed three-helix heterodimer with Cysteine S-sulfate at the N2 position of the alpha-helix | | 分子名称: | 'Cys-N2-SO3-' Strand, 'Positive' Strand, SULFATE ION | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | 登録日 | 2021-01-06 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
5L33
 
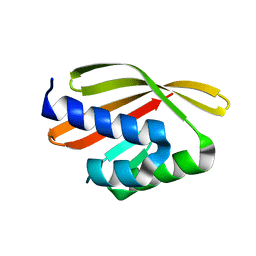 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | denovo NTF2 | | 著者 | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-08-03 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
2CW1
 
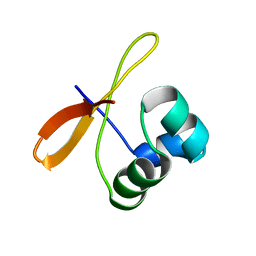 | | Solution structure of the de novo-designed lambda Cro fold protein | | 分子名称: | SN4m | | 著者 | Isogai, Y, Ito, Y, Ikeya, T, Shiro, Y, Ota, M. | | 登録日 | 2005-06-15 | | 公開日 | 2005-12-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of lambda Cro fold: solution structure of a monomeric variant of the de novo protein.
J.Mol.Biol., 354, 2005
|
|
8CCR
 
 | |
3PBJ
 
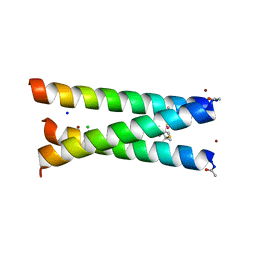 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | 分子名称: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | 著者 | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2010-10-20 | | 公開日 | 2011-11-30 | | 最終更新日 | 2022-05-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
6W3W
 
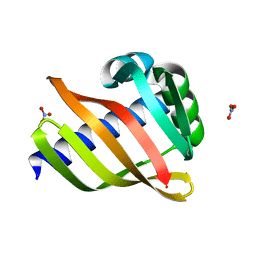 | | An enumerative algorithm for de novo design of proteins with diverse pocket structures | | 分子名称: | DENOVO NTF2, NITRATE ION | | 著者 | Bera, A.K, Basanta, B, Dimaio, F, Sankaran, B, Baker, D. | | 登録日 | 2020-03-09 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | 分子名称: | Computational design, MID1-apo1 | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1E
 
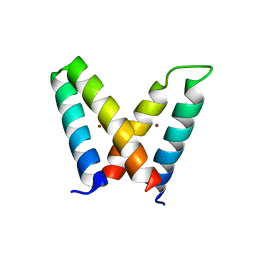 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | 分子名称: | Computational design, MID1-zinc H12E mutant, ZINC ION | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.073 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1C
 
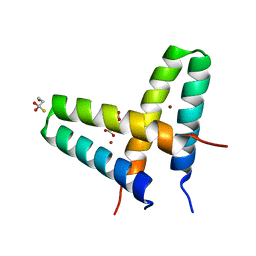 | | Crystal structure of de novo designed MID1-zinc | | 分子名称: | Computational design, MID1-zinc, L(+)-TARTARIC ACID, ... | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.129 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1D
 
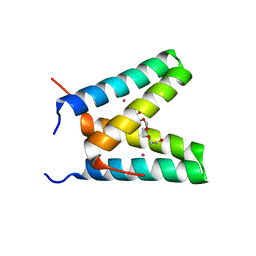 | | Crystal structure of de novo designed MID1-cobalt | | 分子名称: | COBALT (II) ION, Computational design, MID1-cobalt, ... | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.239 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1F
 
 | | Crystal structure of de novo designed MID1-zinc H35E mutant | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Computational design, ... | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.151 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1B
 
 | | Crystal structure of de novo designed MID1-apo2 | | 分子名称: | Computational design, MID1-apo2, GLYCEROL | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
