3CCY
 
 | |
3CD0
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | 分子名称: | (3R,5R)-7-{2-[(4-fluorobenzyl)carbamoyl]-4-(4-fluorophenyl)-1-(1-methylethyl)-1H-imidazol-5-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | 著者 | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | 登録日 | 2008-02-26 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
8SWG
 
 | | RNA duplex bound with GpppA dinucleotide ligand | | 分子名称: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | 著者 | Zhang, W, Dantsu, Y. | | 登録日 | 2023-05-18 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
3CD7
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | 分子名称: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | 著者 | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | 登録日 | 2008-02-26 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
6DXN
 
 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | 分子名称: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | 著者 | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-29 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
8SX5
 
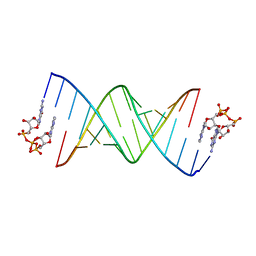 | | GpppA dinucleotide binding to RNA CU template | | 分子名称: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | 著者 | Zhang, W, Dantsu, Y. | | 登録日 | 2023-05-19 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
3CDB
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | 分子名称: | (3R,5R)-7-{3-[(4-carbamoylphenyl)sulfamoyl]-4,5-bis(4-fluorophenyl)-2-(1-methylethyl)-1H-pyrrol-1-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | 著者 | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | 登録日 | 2008-02-26 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CDH
 
 | | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi DSS-3 | | 分子名称: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | 著者 | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-26 | | 公開日 | 2008-03-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi.
To be Published
|
|
3CDL
 
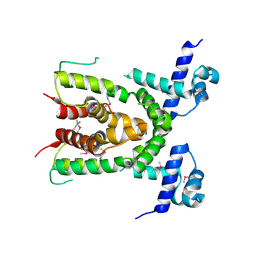 | |
5K26
 
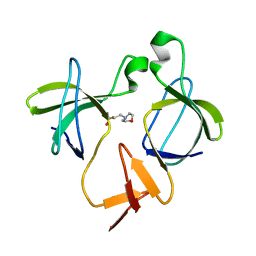 | |
7SF3
 
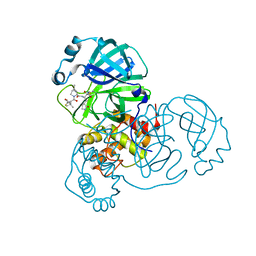 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006m | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-02 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
3C9Z
 
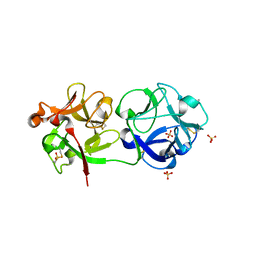 | | Sambucus nigra agglutinin II (SNA-II), tetragonal crystal form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Maveyraud, L, Guillet, V, Mourey, L. | | 登録日 | 2008-02-19 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
5K2P
 
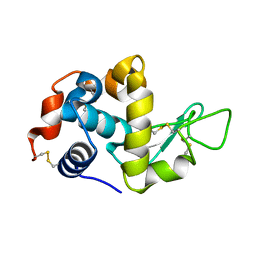 | |
3CDN
 
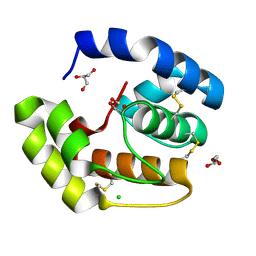 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 4.0 | | 分子名称: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1 | | 著者 | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | 登録日 | 2008-02-27 | | 公開日 | 2008-06-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7SFH
 
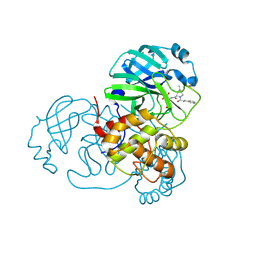 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML102 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-(3-phenylpropanoyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-03 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
3CAP
 
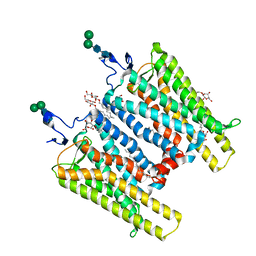 | | Crystal Structure of Native Opsin: the G Protein-Coupled Receptor Rhodopsin in its Ligand-free State | | 分子名称: | 2-O-octyl-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | 著者 | Park, J.H, Scheerer, P, Hofmann, K.P, Choe, H.-W, Ernst, O.P. | | 登録日 | 2008-02-20 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the ligand-free G-protein-coupled receptor opsin
Nature, 454, 2008
|
|
7SGH
 
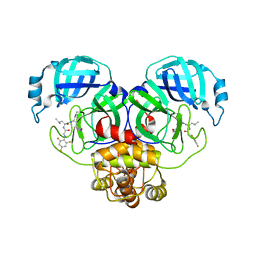 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML124N | | 分子名称: | (S)-N-((S)-1-imino-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-4-methyl-2-(2-((2,4,6-trifluorophenyl)amino)acetamido)pentanamide, 3C-like proteinase | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-05 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
3CB4
 
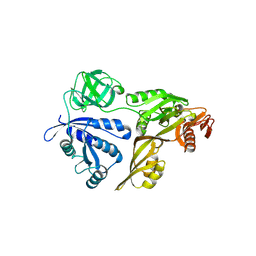 | | The Crystal Structure of LepA | | 分子名称: | GTP-binding protein lepA | | 著者 | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | 登録日 | 2008-02-21 | | 公開日 | 2008-03-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CE4
 
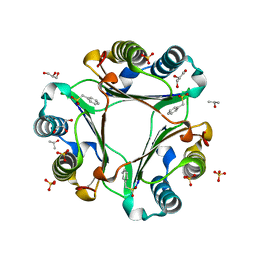 | |
3CAD
 
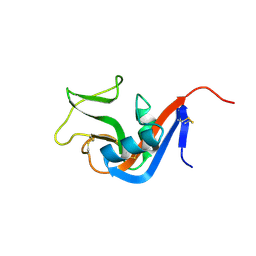 | | Crystal structure of Natural Killer Cell Receptor, Ly49G | | 分子名称: | Lectin-related NK cell receptor LY49G1 | | 著者 | Cho, S. | | 登録日 | 2008-02-19 | | 公開日 | 2008-04-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Architecture of the Major Histocompatibility Complex Class I-binding Site of Ly49 Natural Killer Cell Receptors.
J.Biol.Chem., 283, 2008
|
|
3CAM
 
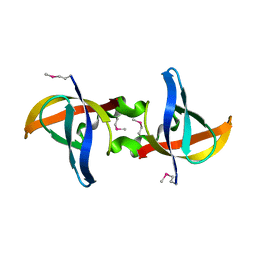 | |
3CAZ
 
 | | Crystal structure of a BAR protein from Galdieria sulphuraria | | 分子名称: | BAR protein | | 著者 | McCoy, J.G, Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2008-02-20 | | 公開日 | 2008-03-04 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.344 Å) | | 主引用文献 | Crystal structure of a BAR protein from Galdieria sulphuraria.
To be Published
|
|
3CB9
 
 | |
8SXL
 
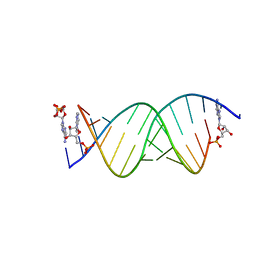 | | RNA UU template binding to AMP monomer | | 分子名称: | ADENOSINE MONOPHOSPHATE, RNA (5'-R(*(TLN)P*(TLN)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | 著者 | Zhang, W, Dantsu, Y. | | 登録日 | 2023-05-22 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
3CBW
 
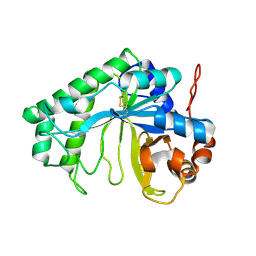 | | Crystal structure of the YdhT protein from Bacillus subtilis | | 分子名称: | CITRIC ACID, YdhT protein | | 著者 | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-23 | | 公開日 | 2008-03-11 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.269 Å) | | 主引用文献 | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
