8ESC
 
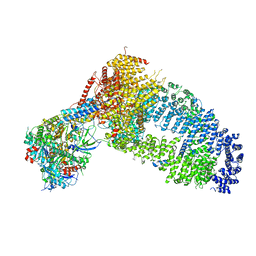 | | Structure of the Yeast NuA4 Histone Acetyltransferase Complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | 著者 | Patel, A.B, Zukin, S.A, Nogales, E. | | 登録日 | 2022-10-13 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure and flexibility of the yeast NuA4 histone acetyltransferase complex.
Elife, 11, 2022
|
|
5DI7
 
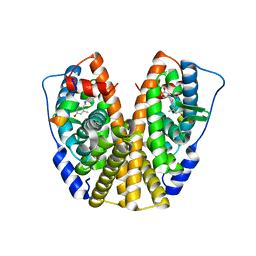 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with an methyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol | | 分子名称: | (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-08-31 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.241 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6ZSP
 
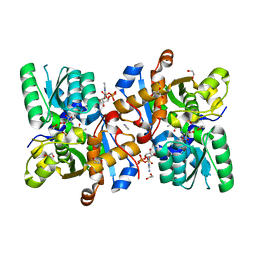 | | Human serine racemase bound to ATP and malonate. | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | 著者 | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | 登録日 | 2020-07-16 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
4WED
 
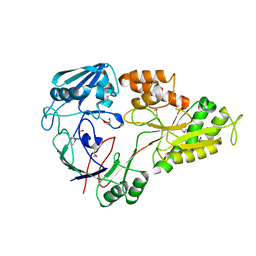 | | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti | | 分子名称: | ABC transporter, periplasmic solute-binding protein, FORMIC ACID, ... | | 著者 | Shabalin, I.G, Otwinowski, Z, Bacal, P, Cymborowski, M.T, Handing, K.B, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-09-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti
to be published
|
|
7SCG
 
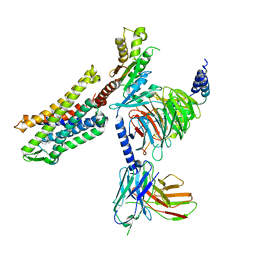 | | FH210 bound Mu Opioid Receptor-Gi Protein Complex | | 分子名称: | (2E)-N-[(2S)-2-(dimethylamino)-3-(4-hydroxyphenyl)propyl]-3-(naphthalen-1-yl)prop-2-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, H, Kobilka, B. | | 登録日 | 2021-09-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3GLY
 
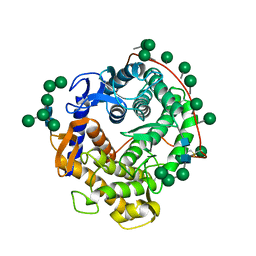 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | 分子名称: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | 登録日 | 1994-06-03 | | 公開日 | 1994-11-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
5HLS
 
 | | Crystal structure of the first bromodomain of human BRD4 bound to CPI-0610 | | 分子名称: | Bromodomain-containing protein 4, CPI-0610 | | 著者 | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | 登録日 | 2016-01-15 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.182 Å) | | 主引用文献 | Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials.
J.Med.Chem., 59, 2016
|
|
8Q54
 
 | |
4WAL
 
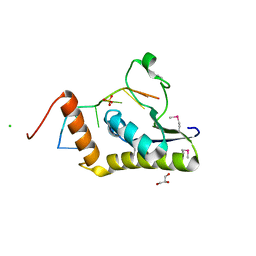 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | 分子名称: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | 著者 | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | 登録日 | 2014-08-29 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
7A9X
 
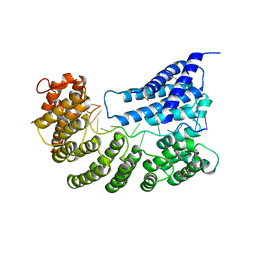 | | Structure of yeast Rmd9p in complex with 16nt target RNA | | 分子名称: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | 著者 | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | 登録日 | 2020-09-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
107L
 
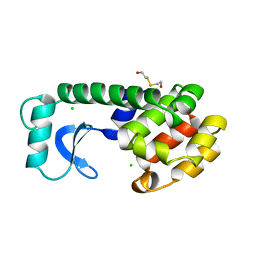 | |
4OUR
 
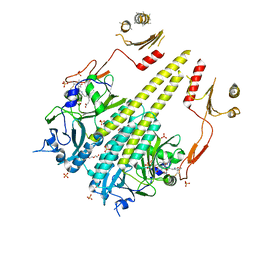 | | Crystal structure of Arabidopsis thaliana phytochrome B photosensory module | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Phytochrome B, ... | | 著者 | Sethe Burgie, E, Bussell, A.N, Walker, J.M, Dubiel, K, Vierstra, R.D. | | 登録日 | 2014-02-18 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
108L
 
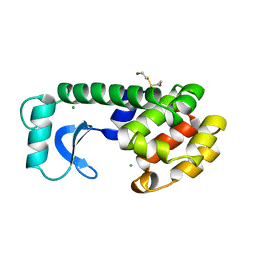 | |
109L
 
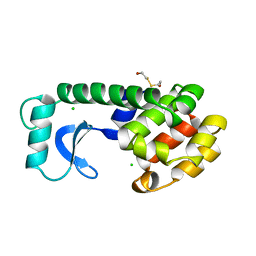 | |
4OVT
 
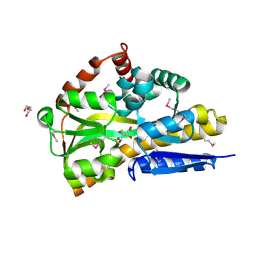 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTERIUM ANTHROPI (Oant_3902), TARGET EFI-510153, WITH BOUND L-FUCONATE | | 分子名称: | 6-deoxy-L-galactonic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-14 | | 公開日 | 2014-01-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
8P8X
 
 | |
6LIX
 
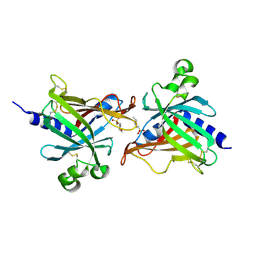 | | CRL Protein of Arabidopsis | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | 著者 | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | 登録日 | 2019-12-13 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-12-02 | | 実験手法 | X-RAY DIFFRACTION (2.385 Å) | | 主引用文献 | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
112L
 
 | |
4ZN9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | 分子名称: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-05-04 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.215 Å) | | 主引用文献 | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
110L
 
 | |
7UQC
 
 | |
111L
 
 | |
113L
 
 | |
5KG9
 
 | |
4UW5
 
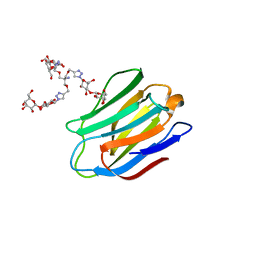 | | Human galectin-7 in complex with a galactose based dendron D2-2. | | 分子名称: | DENDRON D2-1, HUMAN GALECTIN-7 | | 著者 | Ramaswamy, S, Sleiman, M.H, Masuyer, G, Arbez-Gindre, C, Micha-Screttas, M, Calogeropoulou, T, Steele, B.R, Acharya, K.R. | | 登録日 | 2014-08-08 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural Basis of Multivalent Galactose-Based Dendrimer Recognition by Human Galectin-7.
FEBS J., 282, 2015
|
|
