8WHL
 
 | | Crystal structure of CLASP2 in complex with CENP-E | | 分子名称: | ACETATE ION, CLIP-associating protein 2, Centromere-associated protein E, ... | | 著者 | Jia, X, Wei, Z. | | 登録日 | 2023-09-23 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHI
 
 | |
8WHH
 
 | |
8WHM
 
 | | Crystal structure of the ELKS2/LL5beta complex | | 分子名称: | ERC protein 2, Pleckstrin homology-like domain family B member 2 | | 著者 | Jia, X, Wei, Z. | | 登録日 | 2023-09-23 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8IZF
 
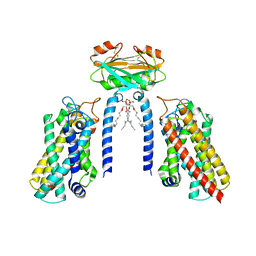 | | Cryo-EM structure of the Lac1-Lip1 (Lip1-S74F) complex | | 分子名称: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1 | | 著者 | Xie, T, Fang, Q, Gong, X. | | 登録日 | 2023-04-07 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8IZD
 
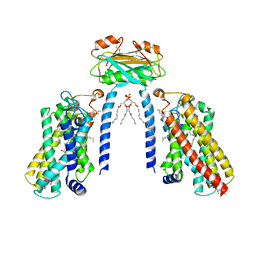 | | Cryo-EM structure of the C26-CoA-bound Lac1-Lip1 complex | | 分子名称: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1, ... | | 著者 | Xie, T, Fang, Q, Gong, X. | | 登録日 | 2023-04-07 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8VYE
 
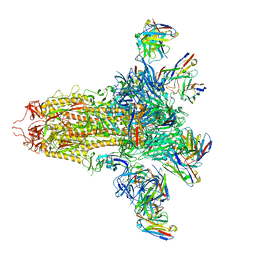 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
7RL3
 
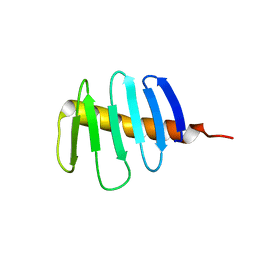 | | Structure of Drosophila melanogaster Plk4 PB3 | | 分子名称: | Serine/threonine-protein kinase PLK4 | | 著者 | Slep, K.C, Slevin, L.K. | | 登録日 | 2021-07-23 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Polo-like kinase 4 homodimerization and condensate formation regulate its own protein levels but are not required for centriole assembly.
Mol.Biol.Cell, 34, 2023
|
|
8VYF
 
 | | SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8WKP
 
 | |
1RK5
 
 | | The D-aminoacylase mutant D366A in complex with 100mM CuCl2 | | 分子名称: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | 著者 | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | 登録日 | 2003-11-20 | | 公開日 | 2004-04-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1RJP
 
 | | Crystal structure of D-aminoacylase in complex with 100mM CuCl2 | | 分子名称: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | 著者 | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | 登録日 | 2003-11-20 | | 公開日 | 2004-04-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
8W22
 
 | | Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) | | 分子名称: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | 著者 | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | 登録日 | 2024-02-19 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
8W20
 
 | | Umb1 umbrella toxin particle | | 分子名称: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | 著者 | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | 登録日 | 2024-02-19 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
6CBI
 
 | | PCNA in complex with inhibitor | | 分子名称: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | 著者 | Bruning, J.B, Wegener, K.L. | | 登録日 | 2018-02-03 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
8OOD
 
 | |
6CCD
 
 | |
6CF6
 
 | | RNF146 TBM-Tankyrase ARC2-3 complex | | 分子名称: | RNF146, Tankyrase-1 | | 著者 | Da Rosa, P.A, Xu, W. | | 登録日 | 2018-02-13 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.
Protein Sci., 27, 2018
|
|
7S1T
 
 | | Structure of the human POT1-TPP1 complex | | 分子名称: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | 著者 | Aramburu, T, Skordalakes, E. | | 登録日 | 2021-09-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | POT1-TPP1 binding stabilizes POT1, promoting efficient telomere maintenance.
Comput Struct Biotechnol J, 20, 2022
|
|
8P8P
 
 | | Crystal structure of human Histidine Triad Nucleotide-Binding Protein 1 in complex with 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl Ethenoadenosine | | 分子名称: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[2-(1~{H}-indol-3-yl)ethyl]carbamate | | 著者 | Dolot, R.M, Dillenburg, M, Wagner, C.R. | | 登録日 | 2023-06-02 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Novel inhibitors for hHINT1 protein
To Be Published
|
|
8PA9
 
 | |
8PAI
 
 | |
8PA6
 
 | |
8PAF
 
 | |
8RJW
 
 | |
