4C6B
 
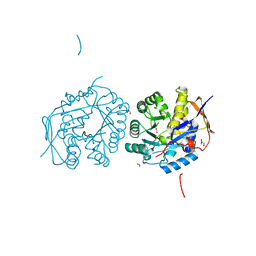 | | Crystal structure of the dihydroorotase domain of human CAD with incomplete active site, obtained recombinantly from E. coli. | | 分子名称: | CAD PROTEIN, FORMIC ACID, GLYCEROL | | 著者 | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.656 Å) | | 主引用文献 | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6P
 
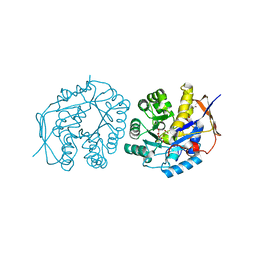 | | Crystal structure of the dihydroorotase domain of human CAD C1613S mutant in apo-form at pH 7.0 | | 分子名称: | CAD PROTEIN, FORMIC ACID, ZINC ION | | 著者 | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.518 Å) | | 主引用文献 | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6Q
 
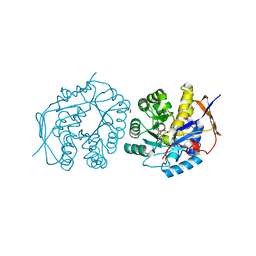 | | Crystal structure of the dihydroorotase domain of human CAD C1613S mutant bound to substrate at pH 7.0 | | 分子名称: | CAD PROTEIN, FORMIC ACID, N-CARBAMOYL-L-ASPARTATE, ... | | 著者 | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.659 Å) | | 主引用文献 | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6J
 
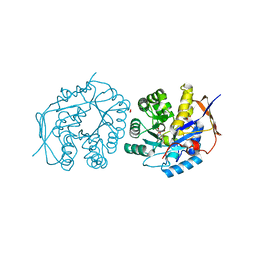 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.5 | | 分子名称: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | 著者 | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.299 Å) | | 主引用文献 | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
1DY3
 
 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | 分子名称: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | 登録日 | 2000-01-21 | | 公開日 | 2000-08-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1DJ2
 
 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | 分子名称: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | 登録日 | 1999-12-01 | | 公開日 | 2000-03-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
3SNV
 
 | |
1CIB
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH GDP, IMP, HADACIDIN, AND NO3 | | 分子名称: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | 著者 | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | 登録日 | 1999-03-31 | | 公開日 | 2000-04-05 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
3V1C
 
 | | Crystal structure of de novo designed MID1-zinc | | 分子名称: | Computational design, MID1-zinc, L(+)-TARTARIC ACID, ... | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.129 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1F
 
 | | Crystal structure of de novo designed MID1-zinc H35E mutant | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Computational design, ... | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.151 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1D
 
 | | Crystal structure of de novo designed MID1-cobalt | | 分子名称: | COBALT (II) ION, Computational design, MID1-cobalt, ... | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.239 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | 著者 | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | 登録日 | 2000-02-22 | | 公開日 | 2001-02-22 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
3V1B
 
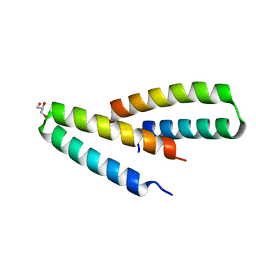 | | Crystal structure of de novo designed MID1-apo2 | | 分子名称: | Computational design, MID1-apo2, GLYCEROL | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
1EZ1
 
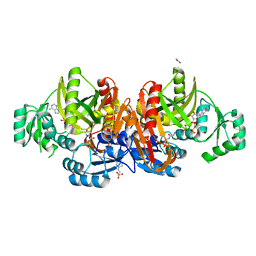 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG, AMPPNP, AND GAR | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETATE ION, GLYCINAMIDE RIBONUCLEOTIDE, ... | | 著者 | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | 登録日 | 2000-05-09 | | 公開日 | 2000-08-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | 著者 | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | 登録日 | 2000-02-25 | | 公開日 | 2001-02-25 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1EYZ
 
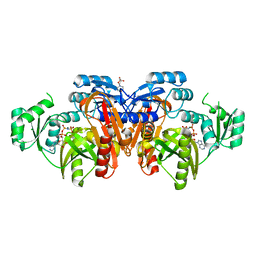 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG AND AMPPNP | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | 登録日 | 2000-05-09 | | 公開日 | 2000-08-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
4IH3
 
 | |
3FGT
 
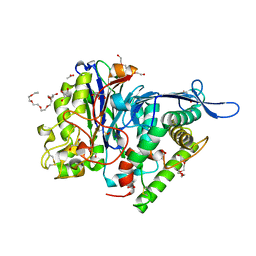 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | 分子名称: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lakomek, K, Dickmanns, A, Ficner, R. | | 登録日 | 2008-12-08 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
6C2V
 
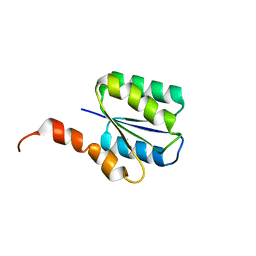 | | Solution structure of a phosphate-loop protein | | 分子名称: | phosphate-loop protein | | 著者 | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | 登録日 | 2018-01-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C2U
 
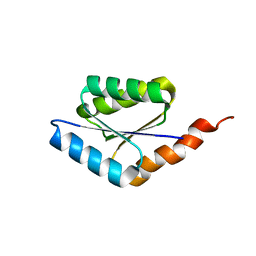 | | Solution structure of a phosphate-loop protein | | 分子名称: | phosphate-loop protein | | 著者 | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | 登録日 | 2018-01-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4J9T
 
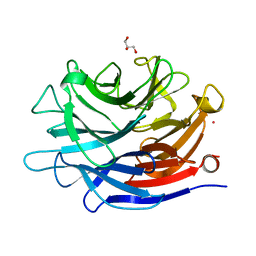 | | Crystal structure of a putative, de novo designed unnatural amino acid dependent metalloprotein, northeast structural genomics consortium target OR61 | | 分子名称: | ARSENIC, GLYCEROL, designed unnatural amino acid dependent metalloprotein | | 著者 | Forouhar, F, Lew, S, Seetharaman, J, Mills, J.H, Khare, S.D, Everett, J.K, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-02-17 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
3FGW
 
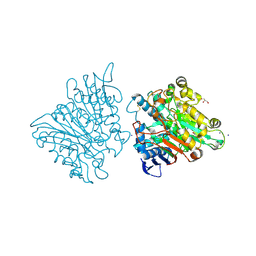 | | One chain form of the 66.3 kDa protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | 著者 | Lakomek, K, Dickmanns, A, Ficner, R. | | 登録日 | 2008-12-08 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGR
 
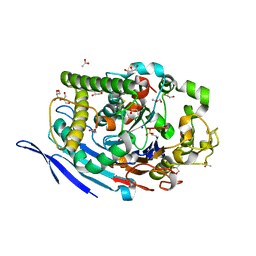 | | Two chain form of the 66.3 kDa protein at 1.8 Angstroem | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Lakomek, K, Dickmanns, A, Ficner, R. | | 登録日 | 2008-12-08 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
6WHO
 
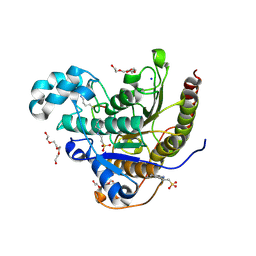 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
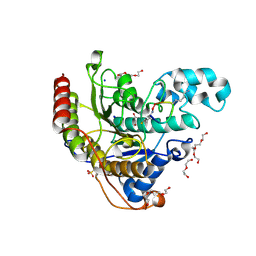 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
