8HY4
 
 | |
8I5Q
 
 | |
8I5S
 
 | | Crystal structure of TxGH116 D593N acid/base mutant from Thermoanaerobacterium xylanolyticum with 2-deoxy-2-fluoroglucoside | | 分子名称: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | 著者 | Pengthaisong, S, Ketudat Cairns, J.R. | | 登録日 | 2023-01-26 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Reaction Mechanism of Glycoside Hydrolase Family 116 Utilizes Perpendicular Protonation.
Acs Catalysis, 13, 2023
|
|
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | 登録日 | 2023-02-14 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | 登録日 | 2023-02-14 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8I5R
 
 | |
2YQ4
 
 | |
1Q3U
 
 | | Crystal structure of a wild-type Cre recombinase-loxP synapse: pre-cleavage complex | | 分子名称: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | 著者 | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | 登録日 | 2003-08-01 | | 公開日 | 2003-09-16 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
8I5T
 
 | |
7JFM
 
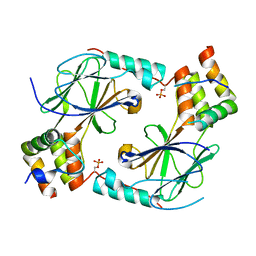 | |
8I5U
 
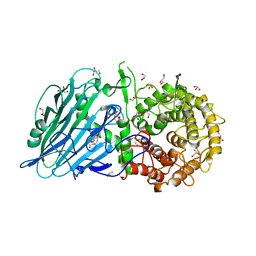 | |
8IBR
 
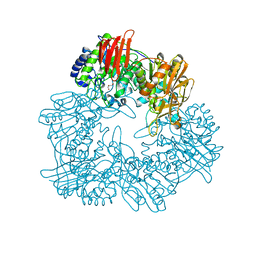 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis in complex with glycerol | | 分子名称: | Beta-galactosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBT
 
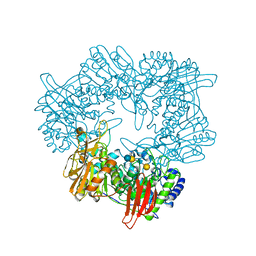 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E318S mutant in complex with lacto-N-tetraose | | 分子名称: | Beta-galactosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBS
 
 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E160A/E318A mutant in complex with galactose | | 分子名称: | Beta-galactosidase, alpha-D-galactopyranose | | 著者 | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
7K4Q
 
 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4V
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K95
 
 | | Crystal structure of human CPSF30 in complex with hFip1 | | 分子名称: | Isoform 2 of Cleavage and polyadenylation specificity factor subunit 4, Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | 著者 | Hamilton, K, Tong, L. | | 登録日 | 2020-09-28 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanism for the interaction between human CPSF30 and hFip1.
Genes Dev., 34, 2020
|
|
7JPV
 
 | | Rabbit Cav1.1 in the presence of 1 micromolar (S)-(-)-Bay K8644 in nanodiscs at 3.4 Angstrom resolution | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, ... | | 著者 | Yan, N, Gao, S. | | 登録日 | 2020-08-10 | | 公開日 | 2020-11-18 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7JRG
 
 | | Plant Mitochondrial complex III2 from Vigna radiata | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Alpha-MPP, ... | | 著者 | Maldonado, M, Letts, J.A. | | 登録日 | 2020-08-12 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
3B4Y
 
 | | FGD1 (Rv0407) from Mycobacterium tuberculosis | | 分子名称: | CITRATE ANION, COENZYME F420, PROBABLE F420-DEPENDENT GLUCOSE-6-PHOSPHATE DEHYDROGENASE FGD1 | | 著者 | Bashiri, G, Squire, C.J, Moreland, N.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2007-10-25 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding
J.Biol.Chem., 283, 2008
|
|
8HYJ
 
 | |
7JX9
 
 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | 分子名称: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | 著者 | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | 登録日 | 2020-08-26 | | 公開日 | 2021-01-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
7JY6
 
 | |
7K1J
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | 著者 | Chen, X, Gellert, M, Yang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
