4JGG
 
 | |
7ZMJ
 
 | | SFX structure of dye-type peroxidase DtpB R243A variant in the ferric state | | 分子名称: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Putative dye-decolorizing peroxidase (DyP), ... | | 著者 | Lucic, M, Worrall, J.A.R, Hough, M.A, Shilova, A, Axford, D.A, Owen, R.L, Tosha, T, Sugimoto, H, Owada, S. | | 登録日 | 2022-04-19 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB.
Acs Catalysis, 12, 2022
|
|
3QMP
 
 | |
8OMC
 
 | |
8OLP
 
 | |
8OQY
 
 | | Structure of apo form of human gamma-secretase PSEN1 APH-1B isoform reconstituted into lipid nanodisc | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Odorcic, I, Chavez Gutierrez, L, Efremov, R.G. | | 登録日 | 2023-04-12 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Apo and A beta 46-bound gamma-secretase structures provide insights into amyloid-beta processing by the APH-1B isoform.
Nat Commun, 15, 2024
|
|
8OQZ
 
 | | Structure of human gamma-secretase PSEN1 APH-1B isoform reconstituted into lipid nanodisc in complex with Ab46 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Odorcic, I, Chavez Gutierrez, L, Efremov, R.G. | | 登録日 | 2023-04-12 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Apo and A beta 46-bound gamma-secretase structures provide insights into amyloid-beta processing by the APH-1B isoform.
Nat Commun, 15, 2024
|
|
8PUQ
 
 | | MetHemoglobin structure from serial synchrotron crystallography with fixed target | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bjelcic, M, Sigfridsson Clauss, K, Aurelius, O, Milas, M, Nan, J, Ursby, T. | | 登録日 | 2023-07-17 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8PUR
 
 | | DexyHemoglobin structure from serial synchrotron crystallography with fixed target | | 分子名称: | Hemoglobin subunit beta, Hemoglobin, alpha 1, ... | | 著者 | Bjelcic, M, Sigfridsson Clauss, K, Aurelius, O, Milas, M, Nan, J, Ursby, T. | | 登録日 | 2023-07-17 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8PWS
 
 | | Dye-decolourising peroxidase DtpB (56 kGy) | | 分子名称: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Lucic, M, Worrall, J.A.R, Hough, M.A, Owen, R.L, Strange, R.W. | | 登録日 | 2023-07-21 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Dye-decolourising peroxidase DtpB (56 kGy)
To Be Published
|
|
8PXP
 
 | |
8PWY
 
 | | Dye-decolourising peroxidase DtpB (112 kGy) | | 分子名称: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Lucic, M, Worrall, J.A.R, Hough, M.A, Owen, R.L, Strange, R.W. | | 登録日 | 2023-07-21 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Dye-decolourising peroxidase DtpB (56 kGy)
To Be Published
|
|
7LK3
 
 | | Crystal structure of untwinned human GABARAPL2 | | 分子名称: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 2 | | 著者 | Scicluna, K, Dewson, G, Czabotar, P.E, Birkinshaw, R.W. | | 登録日 | 2021-02-01 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A new crystal form of GABARAPL2.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4B5L
 
 | |
4P07
 
 | |
4P04
 
 | |
4P06
 
 | |
4RHB
 
 | |
4RH8
 
 | |
4TQP
 
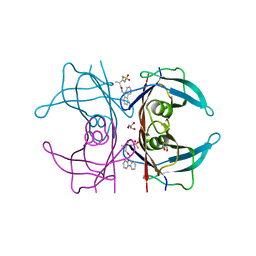 | | Human transthyretin (TTR) complexed with (R)-3-(9H-fluoren-9-ylideneaminooxy)-2-methyl-N-(methylsulfonyl) propionamide in a dual binding mode | | 分子名称: | (2R)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methyl-N-(methylsulfonyl)propanamide, GLYCEROL, Transthyretin | | 著者 | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | 登録日 | 2014-06-11 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
3Q40
 
 | |
4TQ8
 
 | | Dual binding mode for 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid binding to Human transthyretin (TTR) | | 分子名称: | 1,2-ETHANEDIOL, 3-[(9H-fluoren-9-ylideneamino)oxy]propanoic acid, Transthyretin | | 著者 | Ciccone, L, Orlandini, E, Nencetti, S, Rossello, A, Stura, E.A. | | 登録日 | 2014-06-10 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
4TQH
 
 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)ethanoic acid | | 分子名称: | 1,2-ETHANEDIOL, Transthyretin, [(9H-fluoren-9-ylideneamino)oxy]acetic acid | | 著者 | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | 登録日 | 2014-06-11 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.511 Å) | | 主引用文献 | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
3Q5G
 
 | |
2X0A
 
 | |
