4ZBI
 
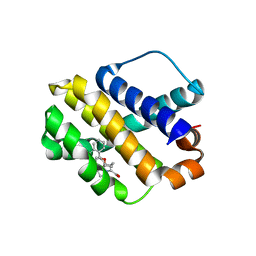 | | Mcl-1 complexed with small molecules | | 分子名称: | 1-[3-(naphthalen-1-yloxy)propyl]-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Zhao, B. | | 登録日 | 2015-04-14 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
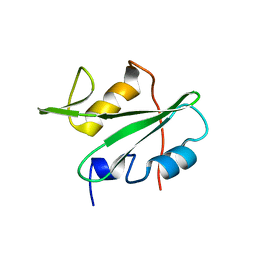
3EAZ
 
 | |
3GIN
 
 | |
3FHO
 
 | | Structure of S. pombe Dbp5 | | 分子名称: | ATP-dependent RNA helicase dbp5 | | 著者 | Cheng, Z, Song, H. | | 登録日 | 2008-12-09 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
4A55
 
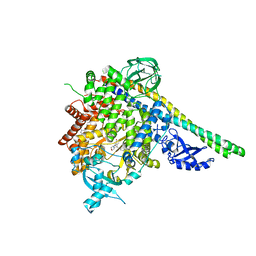 | | Crystal structure of p110alpha in complex with iSH2 of p85alpha and the inhibitor PIK-108 | | 分子名称: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM, PIK-108 | | 著者 | Hon, W.-C, Berndt, A, Williams, R.L. | | 登録日 | 2011-10-24 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Regulation of Lipid Binding Underlies the Activation Mechanism of Class Ia Pi3-Kinases.
Oncogene, 31, 2012
|
|
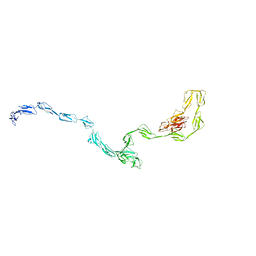
3GAU
 
 | |
8EZD
 
 | |
8EZE
 
 | |
3GGW
 
 | |
3GAV
 
 | |
3GAW
 
 | |
3HQB
 
 | |
3HQA
 
 | |
4AGW
 
 | | Discovery of a small molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromet hyl)phenyl}benzamide, GLYCEROL, ... | | 著者 | Weisberg, E, Choi, H.G, Seeliger, M, Gray, N, Griffin, J.D. | | 登録日 | 2012-02-01 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of a Small-Molecule Type II Inhibitor of Wild-Type and Gatekeeper Mutants of Bcr-Abl, Pdgfralpha, Kit, and Src Kinases: Novel Type II Inhibitor of Gatekeeper Mutants.
Blood, 115, 2010
|
|
3EAC
 
 | |
3LNX
 
 | | Second PDZ domain from human PTP1E | | 分子名称: | IODIDE ION, THIOCYANATE ION, Tyrosine-protein phosphatase non-receptor type 13 | | 著者 | Zhang, J, Chang, A, Ke, H, Phillips Jr, G.N, Lee, A.L, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2010-02-03 | | 公開日 | 2010-02-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.642 Å) | | 主引用文献 | Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E.
Biochemistry, 49, 2010
|
|
3E0U
 
 | | Crystal structure of T. cruzi GPX1 | | 分子名称: | AMMONIUM ION, GLYCEROL, Glutathione peroxidase | | 著者 | Patel, S.H, Hussain, S, Harris, R, Driscoll, P, Djordjevic, S. | | 登録日 | 2008-08-01 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of Trypanosoma cruzi GPXI (glutathione peroxidase-like enzyme I).
Biochem.J., 425, 2010
|
|
6R3M
 
 | | Family 11 Carbohydrate-Binding Module from Clostridium thermocellum in complex with beta-1,3-1,4-mixed-linked tetrasaccharide | | 分子名称: | ACETATE ION, CALCIUM ION, Endoglucanase H, ... | | 著者 | Ribeiro, D.O, Carvalho, A.L. | | 登録日 | 2019-03-20 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular basis for the preferential recognition of beta 1,3-1,4-glucans by the family 11 carbohydrate-binding module from Clostridium thermocellum.
Febs J., 287, 2020
|
|
6R4W
 
 | |
6R56
 
 | | Crystal structure of PPEP-1(K101E/E184K) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | 著者 | Pichlo, C, Baumann, U. | | 登録日 | 2019-03-24 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6R4Z
 
 | |
6R59
 
 | |
6LF1
 
 | | SeviL, a GM1b/asialo-GM1 binding lectin | | 分子名称: | CHLORIDE ION, SeviL | | 著者 | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2019-11-28 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
5HS7
 
 | |
6R36
 
 | | T. brucei farnesyl pyrophosphate synthase (FPPS) | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Farnesyl pyrophosphate synthase | | 著者 | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | 登録日 | 2019-03-19 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Fragment-Based Discovery of Non-bisphosphonate Binders of Trypanosoma brucei Farnesyl Pyrophosphate Synthase.
Chembiochem, 21, 2020
|
|
