1SK4
 
 | | crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | 分子名称: | Peptidoglycan recognition protein I-alpha, SODIUM ION | | 著者 | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | 登録日 | 2004-03-04 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
3SUZ
 
 | |
385D
 
 | |
3A5T
 
 | | Crystal structure of MafG-DNA complex | | 分子名称: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | 著者 | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | 登録日 | 2009-08-11 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 2 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3TWS
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human TERF1 (chimeric peptide) | | 分子名称: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, ... | | 著者 | Guettler, S, Sicheri, F. | | 登録日 | 2011-09-22 | | 公開日 | 2011-12-07 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3ONG
 
 | | Crystal structure of UBA2ufd-Ubc9: insights into E1-E2 interactions in Sumo pathways | | 分子名称: | SUMO-conjugating enzyme UBC9, Ubiquitin-activating enzyme E1-like | | 著者 | Wang, J, Taherbhoy, A.M, Hunt, H.W, Seyedin, S.N, Miller, D.W, Huang, D.T, Schulman, B.A. | | 登録日 | 2010-08-28 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.
Plos One, 5, 2010
|
|
4JCJ
 
 | | Crystal structure of Isl1 LIM domains with Ldb1 LIM-interaction domain | | 分子名称: | Insulin gene enhancer protein ISL-1,LIM domain-binding protein 1, ZINC ION | | 著者 | Gadd, M.S, Jacques, D.A, Guss, J.M, Matthews, J.M. | | 登録日 | 2013-02-21 | | 公開日 | 2013-06-19 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A structural basis for the regulation of the LIM-homeodomain protein islet 1 (Isl1) by intra- and intermolecular interactions.
J.Biol.Chem., 288, 2013
|
|
3A8N
 
 | | Crystal structure of the Tiam1 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 1 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
4J5Q
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1, apo structure | | 分子名称: | O-acetyl-ADP-ribose deacetylase 1 | | 著者 | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | 登録日 | 2013-02-09 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
1GPR
 
 | |
3P0P
 
 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | 分子名称: | 2-[4-(4-fluorophenyl)piperazin-1-yl]-6-methylpyrimidin-4(3H)-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-09-29 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
4J21
 
 | | Tankyrase 2 in complex with 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one | | 分子名称: | 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | 登録日 | 2013-02-04 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
3ONJ
 
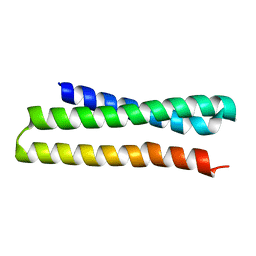 | |
4J3L
 
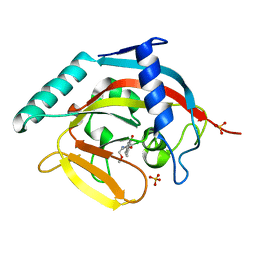 | | Tankyrase 2 in complex with 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide | | 分子名称: | 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide, SULFATE ION, Tankyrase-2, ... | | 著者 | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | 登録日 | 2013-02-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
1ILY
 
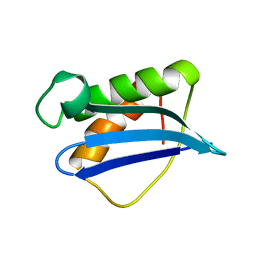 | | Solution Structure of Ribosomal Protein L18 of Thermus thermophilus | | 分子名称: | RIBOSOMAL PROTEIN L18 | | 著者 | Woestenenk, E.A, Gongadze, G.M, Shcherbakov, D.V, Rak, A.V, Garber, M.B, Hard, T, Berglund, H. | | 登録日 | 2001-05-09 | | 公開日 | 2002-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold.
Biochem.J., 363, 2002
|
|
4IWC
 
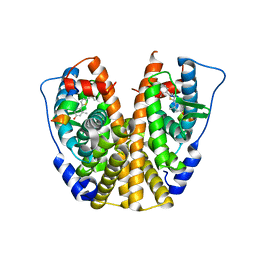 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with a Dynamic Thiophene-derivative | | 分子名称: | 4,4'-thiene-2,5-diylbis(3-methylphenol), Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2013-01-23 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
3PN5
 
 | |
4IWT
 
 | |
4IUU
 
 | |
4IVV
 
 | |
4IW6
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 7b | | 分子名称: | 4-[2-(but-3-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2013-01-23 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
3OTU
 
 | | PDK1 mutant bound to allosteric disulfide fragment activator JS30 | | 分子名称: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(naphthalen-1-ylmethyl)piperazin-1-yl]-4-oxobutane-1-thiol, ... | | 著者 | Sadowsky, J.D, Wells, J.A. | | 登録日 | 2010-09-13 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1013 Å) | | 主引用文献 | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BV3
 
 | | Morpholino pyrrolotriazine P38 Alpha Map Kinase inhibitor compound 2 | | 分子名称: | 5-methyl-4-[(2-methyl-5-{[(3-morpholin-4-ylphenyl)carbonyl]amino}phenyl)amino]-N-[(1S)-1-phenylethyl]pyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | 著者 | Sack, J.S. | | 登録日 | 2008-01-04 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1IFY
 
 | |
