7O6L
 
 | |
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | 分子名称: | ACYLATED CEFTAZIDIME, Beta-lactamase | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-12 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
5T0J
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2016-08-16 | | 公開日 | 2016-10-19 | | 最終更新日 | 2016-11-30 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4WH0
 
 | | YcaC from Pseudomonas aeruginosa with S-mercaptocysteine active site cysteine | | 分子名称: | CHLORIDE ION, Putative hydrolase | | 著者 | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | 登録日 | 2014-09-19 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.563 Å) | | 主引用文献 | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
6UT8
 
 | | Refined half-complex from tetradecameric assembly of Thermococcus gammatolerans McrB AAA+ hexamers with bound McrC | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | 登録日 | 2019-10-29 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
7OD5
 
 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP crystallization | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | 著者 | Gabdulkhakov, A.G, Selikhanov, G.K, Fufina, T.Y, Vasilieva, L.G. | | 登録日 | 2021-04-28 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray structure of the Rhodobacter sphaeroides reaction center with an M197 Phe→His substitution clarifies the properties of the mutant complex.
Iucrj, 9, 2022
|
|
8FM6
 
 | |
6V5E
 
 | | Crystal structure of CTX-M-14 P167S/D240G beta-lactamase | | 分子名称: | Beta-lactamase | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-04 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
3ZIN
 
 | | Gu_alpha_helicase | | 分子名称: | IMPORTIN SUBUNIT ALPHA-2, NUCLEOLAR RNA HELICASE 2 | | 著者 | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | 登録日 | 2013-01-10 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
5T0I
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2016-08-16 | | 公開日 | 2016-10-19 | | 最終更新日 | 2016-11-30 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MIS
 
 | | Native ananain in complex with E-64 | | 分子名称: | Ananain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | 著者 | Yongqing, T, Wilmann, P.G, Pike, R.N, Wijeyewickrema, L.C. | | 登録日 | 2018-09-20 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Determination of the crystal structure and substrate specificity of ananain.
Biochimie, 166, 2019
|
|
6V3C
 
 | |
6V7T
 
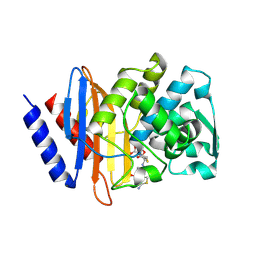 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | 分子名称: | ACYLATED CEFTAZIDIME, Beta-lactamase | | 著者 | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | 登録日 | 2019-12-09 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6VAG
 
 | | Crystal structure of the oligomerization domain of phosphoprotein from parainfluenza virus 5 | | 分子名称: | GLYCEROL, Phosphoprotein | | 著者 | Aggarwal, M, Abdella, R, He, Y, Lamb, R.A. | | 登録日 | 2019-12-17 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5T0G
 
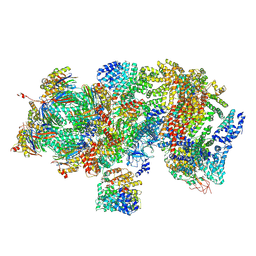 | | Structural basis for dynamic regulation of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2016-08-16 | | 公開日 | 2016-10-19 | | 最終更新日 | 2016-11-30 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3ZIO
 
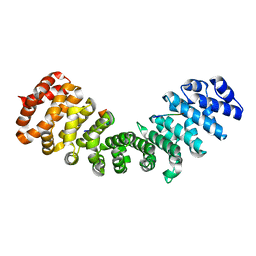 | | minor-site specific NLS (A28) | | 分子名称: | A28NLS, IMPORTIN SUBUNIT ALPHA-2 | | 著者 | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | 登録日 | 2013-01-10 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
8QFD
 
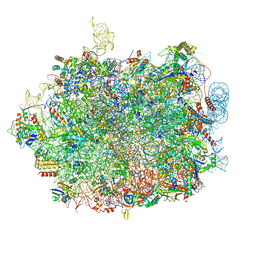 | | UFL1 E3 ligase bound 60S ribosome | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Makhlouf, L, Kulathu, Y, Zeqiraj, E. | | 登録日 | 2023-09-04 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
1G4C
 
 | |
6WA8
 
 | |
5K9N
 
 | | Structural and Mechanistic Analysis of Drosophila melanogaster Polyamine N acetyltransferase, an enzyme that Catalyzes the Formation of N acetylagmatine | | 分子名称: | Polyamine N acetyltransferase | | 著者 | Dempsey, D.R, Nichols, D.A, Battistini, M.R, Pemberton, O, Ospina, S.R, Zhang, X, Carpenter, A.-M, Chen, Y, Merkler, D.J. | | 登録日 | 2016-06-01 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Mechanistic Analysis of Drosophila melanogaster Agmatine N-Acetyltransferase, an Enzyme that Catalyzes the Formation of N-Acetylagmatine.
Sci Rep, 7, 2017
|
|
7OMG
 
 | | Crystal structure of KOD DNA Polymerase in a ternary complex with an Uracil containing template | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | 登録日 | 2021-05-22 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OMB
 
 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | 著者 | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | 登録日 | 2021-05-21 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OM3
 
 | | Crystal structure of KOD DNA Polymerase in a binary complex with Hypoxanthine containing template | | 分子名称: | 1,2-ETHANEDIOL, 21nt Template, BROMIDE ION, ... | | 著者 | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | 登録日 | 2021-05-21 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
6W3Q
 
 | |
6LZJ
 
 | | Aquifex aeolicus MutL ATPase domain complexed with AMPPCP | | 分子名称: | DNA mismatch repair protein MutL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Fukui, K, Izuhara, K, Yano, T. | | 登録日 | 2020-02-19 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.72835565 Å) | | 主引用文献 | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
