3K8D
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | 分子名称: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | 登録日 | 2009-10-14 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
1O67
 
 | |
7ACF
 
 | |
3HRF
 
 | | Crystal structure of Human PDK1 kinase domain in complex with an allosteric activator bound to the PIF-pocket | | 分子名称: | (2Z)-5-(4-chlorophenyl)-3-phenylpent-2-enoic acid, 3-phosphoinositide-dependent protein kinase 1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Hindie, V, Alzari, P.M, Biondi, R.M. | | 登録日 | 2009-06-09 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1.
Nat.Chem.Biol., 5, 2009
|
|
7Y9Y
 
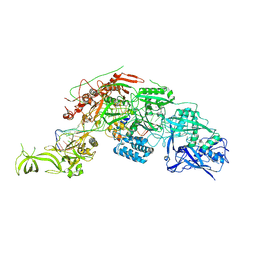 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | 分子名称: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | 著者 | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-06-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
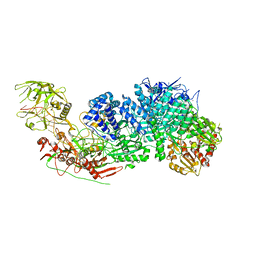 | | Structure of the Cas7-11-Csx29-guide RNA complex | | 分子名称: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | 著者 | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-06-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
4YAZ
 
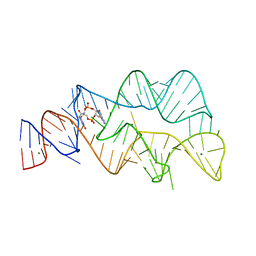 | | 3',3'-cGAMP riboswitch bound with 3',3'-cGAMP | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | 登録日 | 2015-02-18 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
6ZCI
 
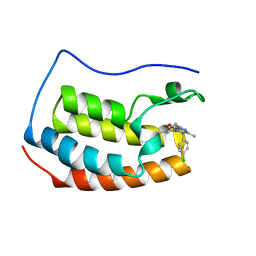 | | Crystal structure of BRD4-BD1 in complex with NVS-BET-1 | | 分子名称: | (4~{R})-4-(4-chlorophenyl)-1-cyclopropyl-5-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)-3-methyl-4~{H}-pyrrolo[3,4-c]pyrazol-6-one, Bromodomain-containing protein 4 | | 著者 | Faller, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.976 Å) | | 主引用文献 | BET bromodomain inhibitors regulate keratinocyte plasticity.
Nat.Chem.Biol., 17, 2021
|
|
4YHX
 
 | |
3HLO
 
 | |
4YB1
 
 | | 20A Mutant c-di-GMP Vc2 Riboswitch bound with 3',3'-cGAMP | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, RNA (91-MER), ... | | 著者 | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | 登録日 | 2015-02-18 | | 公開日 | 2015-04-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
7ZL1
 
 | | PTX3 Pentraxin Domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pentraxin-related protein PTX3 | | 著者 | Noone, D.P, Sharp, T.H. | | 登録日 | 2022-04-13 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1O61
 
 | |
7ZL4
 
 | | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | | 分子名称: | CsuA/B | | 著者 | Pakharukova, N, Malmi, H, Tuittila, M, Paavilainen, S, Ghosal, D, Chang, Y.W, Jensen, G.J, Zavialov, A.V. | | 登録日 | 2022-04-13 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.
Nature, 609, 2022
|
|
6Z5V
 
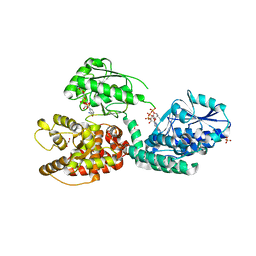 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3-KETODECANOYL-COA IN CROTONASE FOLD AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE IN HAD FOLD | | 分子名称: | 3-KETO-DECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | 登録日 | 2020-05-27 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1NRK
 
 | |
4Z4A
 
 | |
7A54
 
 | |
7ZH0
 
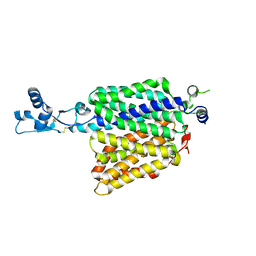 | |
7ZOU
 
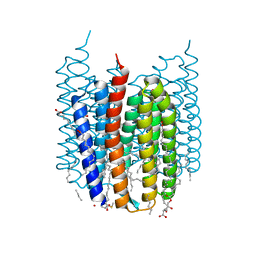 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, ground state | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, EICOSANE, ... | | 著者 | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | 登録日 | 2022-04-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
4Z7U
 
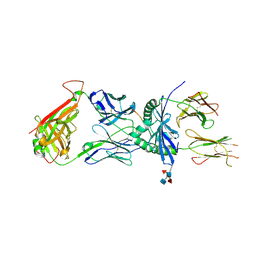 | | S13 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DQ-alpha chain, MHC class II HLA-DQ-beta-1, ... | | 著者 | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | 登録日 | 2015-04-08 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
6ZMT
 
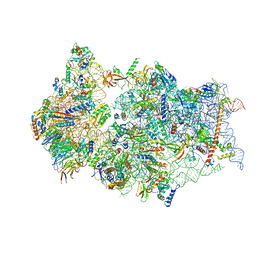 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-03 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP0
 
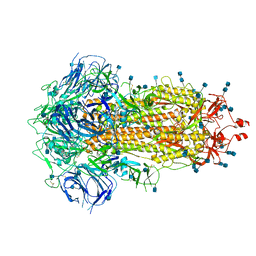 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-06-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7ZOV
 
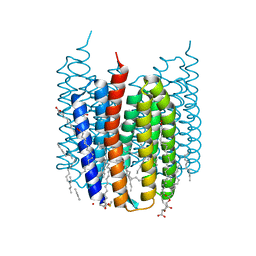 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, K state | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, EICOSANE, ... | | 著者 | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | 登録日 | 2022-04-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
7ZH6
 
 | |
