8YGG
 
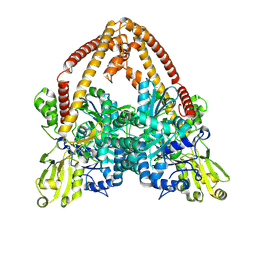 | | pP1192R-apo Closed state | | 分子名称: | DNA topoisomerase 2 | | 著者 | Sun, J.Q, Liu, R.L. | | 登録日 | 2024-02-26 | | 公開日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 2024
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | 著者 | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | 登録日 | 2015-08-26 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
7VZ4
 
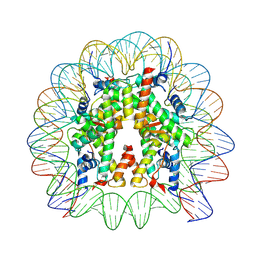 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601L DNA sequence | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Takizawa, Y, Ho, C.-H, Sato, S, Danev, R, Kurumizaka, H. | | 登録日 | 2021-11-15 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (1.89 Å) | | 主引用文献 | Methods for High Resolution Cryo-EM Analyses of Nucleosomes
To Be Published
|
|
1CR0
 
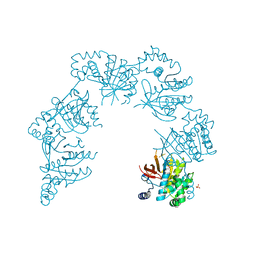 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE4 PROTEIN OF BACTERIOPHAGE T7 | | 分子名称: | DNA PRIMASE/HELICASE, SULFATE ION | | 著者 | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | 登録日 | 1999-08-12 | | 公開日 | 1999-11-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
7LV8
 
 | | Structure of the Marseillevirus nucleosome | | 分子名称: | DNA (123-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | 著者 | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | 登録日 | 2021-02-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8J91
 
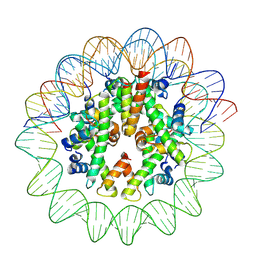 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones | | 分子名称: | DNA (169-MER), HTA13, Histone H2B.6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
1AG3
 
 | |
2RVB
 
 | |
2IZO
 
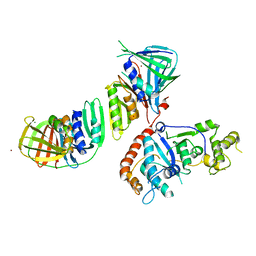 | | Structure of an Archaeal PCNA1-PCNA2-FEN1 Complex | | 分子名称: | DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C, FLAP STRUCTURE-SPECIFIC ENDONUCLEASE, ... | | 著者 | Dore, A.S, Kilkenny, M.L, Roe, S.M, Pearl, L.H. | | 登録日 | 2006-07-25 | | 公開日 | 2006-09-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of an Archaeal PCNA1-PCNA2-Fen1 Complex: Elucidating PCNA Subunit and Client Enzyme Specificity.
Nucleic Acids Res., 34, 2006
|
|
1D42
 
 | |
8D7U
 
 | |
8D7Y
 
 | |
8D7W
 
 | |
8D7X
 
 | |
8CVP
 
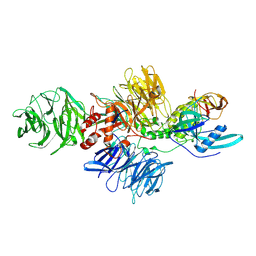 | | Cereblon-DDB1 in the Apo form | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7V
 
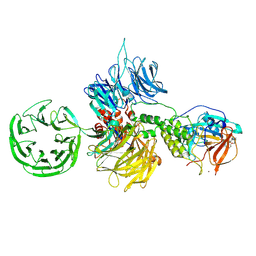 | |
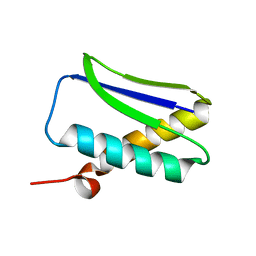
1GHH
 
 | |
1D90
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH I.T MISMATCHED BASE PAIRS | | 分子名称: | DNA (5'-D(*GP*GP*IP*GP*CP*TP*CP*C)-3') | | 著者 | Cruse, W.B.T, Aymani, J, Kennard, O, Brown, T, Jack, A.G.C, Leonard, G.A. | | 登録日 | 1992-10-17 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs.
Nucleic Acids Res., 17, 1989
|
|
1D9H
 
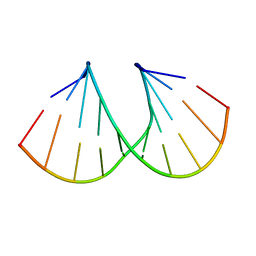 | | Structural origins of the exonuclease resistance of a zwitterionic RNA | | 分子名称: | DNA/RNA (5'-D(*GP*CP*GP*TP*AP)-R(*(U31)P)-D(*AP*CP*GP*C)-3') | | 著者 | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | 登録日 | 1999-10-27 | | 公開日 | 1999-12-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural origins of the exonuclease resistance of a zwitterionic RNA
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4Q0R
 
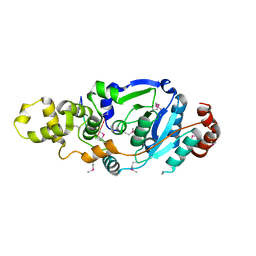 | | The catalytic core of Rad2 (complex I) | | 分子名称: | DNA (5'-D(*CP*TP*GP*AP*GP*TP*CP*AP*GP*AP*GP*CP*AP*AP*A)-3'), DNA repair protein RAD2 | | 著者 | Mietus, M, Nowak, E, Jaciuk, M, Kustosz, P, Nowotny, M. | | 登録日 | 2014-04-02 | | 公開日 | 2014-08-27 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding.
Nucleic Acids Res., 42, 2014
|
|
1MSZ
 
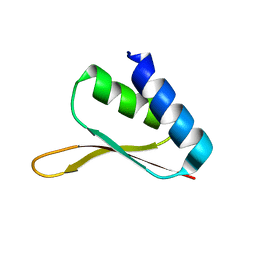 | | Solution structure of the R3H domain from human Smubp-2 | | 分子名称: | DNA-binding protein SMUBP-2 | | 著者 | Liepinsh, E, Leonchiks, A, Sharipo, A, Guignard, L, Otting, G. | | 登録日 | 2002-09-20 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the R3H domain from human Smubp-2
J.Mol.Biol., 326, 2003
|
|
6RR9
 
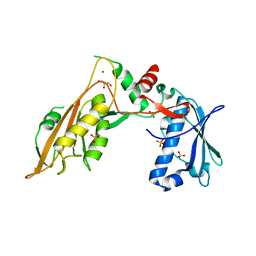 | | DNA/RNA binding protein | | 分子名称: | GLYCEROL, SULFATE ION, Schlafen family member 5, ... | | 著者 | Huber, E, Lammens, K. | | 登録日 | 2019-05-17 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (3.432 Å) | | 主引用文献 | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 2022
|
|
7VBM
 
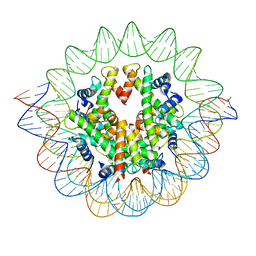 | | The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv | | 分子名称: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | 登録日 | 2021-08-31 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
317D
 
 | |
4A11
 
 | | Structure of the hsDDB1-hsCSA complex | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | 著者 | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | 登録日 | 2011-09-13 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
