5ZU2
 
 | | Effect of mutation (R554A) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Mikami, B, Uchida, H, Doubayashi, D. | | 登録日 | 2018-05-06 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.441 Å) | | 主引用文献 | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
5ZU3
 
 | | Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Formate oxidase, ... | | 著者 | Mikami, B, Uchida, H, Doubayashi, D. | | 登録日 | 2018-05-06 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
1C5H
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | 分子名称: | ENDO-1,4-BETA-XYLANASE | | 著者 | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | 登録日 | 1999-11-24 | | 公開日 | 2000-05-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
1C5I
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | 分子名称: | ENDO-1,4-BETA-XYLANASE, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | 著者 | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | 登録日 | 1999-11-24 | | 公開日 | 2000-05-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
7BSU
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-bound E2BeF state | | 分子名称: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | 著者 | Abe, K, Nishizawa, T, Nakanishi, H. | | 登録日 | 2020-03-31 | | 公開日 | 2020-09-30 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BSW
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdEtn-occluded E2-AlF state | | 分子名称: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | 著者 | Abe, K, Nishizawa, T, Nakanishi, H. | | 登録日 | 2020-03-31 | | 公開日 | 2020-09-30 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | 登録日 | 2021-03-15 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
1CGU
 
 | | CATALYTIC CENTER OF CYCLODEXTRIN GLYCOSYLTRANSFERASE DERIVED FROM X-RAY STRUCTURE ANALYSIS COMBINED WITH SITE-DIRECTED MUTAGENESIS | | 分子名称: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Klein, C, Hollender, J, Bender, H, Schulz, G.E. | | 登録日 | 1992-06-10 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis.
Biochemistry, 31, 1992
|
|
3CPU
 
 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | 分子名称: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | 著者 | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | 登録日 | 1999-06-08 | | 公開日 | 2001-06-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | 著者 | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | 登録日 | 2000-07-26 | | 公開日 | 2001-09-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
3TB4
 
 | | Crystal structure of the ISC domain of VibB | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Liu, S, Zhang, C, Niu, B, Li, N, Liu, M, Wei, T, Zhu, D, Xu, S, Gu, L. | | 登録日 | 2011-08-05 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1YYB
 
 | |
4O4D
 
 | | Crystal Structure of an Inositol hexakisphosphate kinase EhIP6KA in complexed with ATP and Ins(1,4,5)P3 | | 分子名称: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family.
Nat Commun, 5, 2014
|
|
4O4F
 
 | |
4O4C
 
 | |
4O4B
 
 | |
4O4E
 
 | | Crystal Structure of an Inositol hexakisphosphate kinase EhIP6KA in complexed with ATP and Ins(1,3,4,5,6)P5 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate kinase, MAGNESIUM ION, ... | | 著者 | Wang, H, Shears, S.B. | | 登録日 | 2013-12-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family.
Nat Commun, 5, 2014
|
|
3TG2
 
 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | 分子名称: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | 著者 | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | 登録日 | 2011-08-17 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2A7T
 
 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | 分子名称: | Neurotoxin | | 著者 | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | 登録日 | 2005-07-06 | | 公開日 | 2005-07-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
2CPU
 
 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | 分子名称: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | 著者 | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | 登録日 | 1999-06-08 | | 公開日 | 2001-06-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
2CXG
 
 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | 分子名称: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | 著者 | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | 登録日 | 1998-05-08 | | 公開日 | 1998-10-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
2ECH
 
 | |
1L6E
 
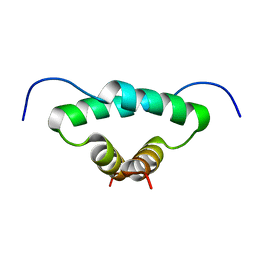 | | Solution structure of the docking and dimerization domain of protein kinase A II-alpha (RIIalpha D/D). Alternatively called the N-terminal dimerization domain of the regulatory subunit of protein kinase A. | | 分子名称: | cAMP-dependent protein kinase Type II-alpha regulatory chain | | 著者 | Morikis, D, Roy, M, Newlon, M.G, Scott, J.D, Jennings, P.A. | | 登録日 | 2002-03-08 | | 公開日 | 2002-04-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Electrostatic properties of the structure of the docking and dimerization domain of protein kinase A IIalpha
Eur.J.Biochem., 269, 2002
|
|
6R0R
 
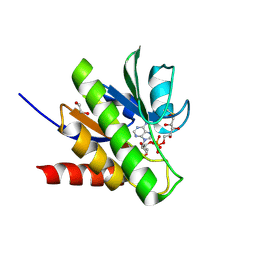 | | Getah virus macro domain in complex with ADPr covalently bond to Cys34 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural polyprotein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S})-2,3,4,5-tetrakis(oxidanyl)pentyl] hydrogen phosphate | | 著者 | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | 登録日 | 2019-03-13 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
1A8M
 
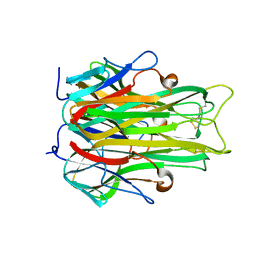 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | 分子名称: | TUMOR NECROSIS FACTOR ALPHA | | 著者 | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | 登録日 | 1998-03-27 | | 公開日 | 1998-06-17 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
