5BKG
 
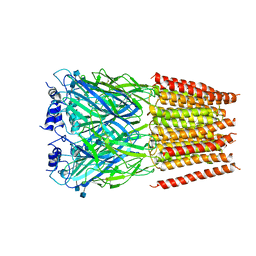 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, glycine bound, (semi)open state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | 著者 | Yu, H, Wang, W. | | 登録日 | 2021-03-19 | | 公開日 | 2021-09-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5BKF
 
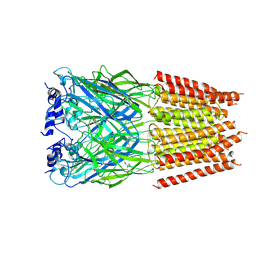 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, Glycine bound, desensitized state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | 著者 | Yu, H, Wang, W. | | 登録日 | 2021-03-19 | | 公開日 | 2021-09-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5BJP
 
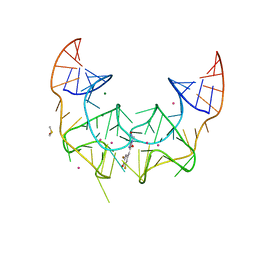 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, iridium hexammine soak | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, DIMETHYL SULFOXIDE, IRIDIUM ION, ... | | 著者 | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | 登録日 | 2016-08-21 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
5BJO
 
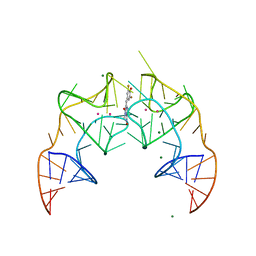 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, site-specific 5-iodo-U | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | 登録日 | 2016-08-21 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
5B61
 
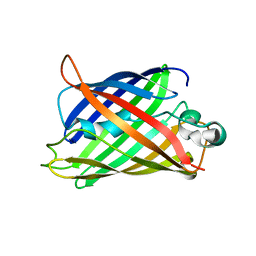 | | Extra-superfolder GFP | | 分子名称: | Green fluorescent protein | | 著者 | Park, H.H, Jang, T.-H, Choi, J.Y. | | 登録日 | 2016-05-24 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.115 Å) | | 主引用文献 | The mechanism of folding robustness revealed by the crystal structure of extra-superfolder GFP.
FEBS Lett., 591, 2017
|
|
5AZC
 
 | |
5AZB
 
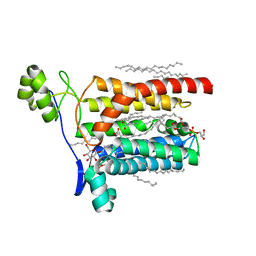 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | 著者 | Zhang, X.C, Mao, G, Zhao, Y. | | 登録日 | 2015-09-30 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
5AQB
 
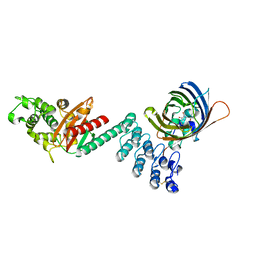 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | 分子名称: | 3G61_DB15V4, GREEN FLUORESCENT PROTEIN | | 著者 | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | 登録日 | 2015-09-21 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | 登録日 | 2015-02-19 | | 公開日 | 2015-05-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
5AB7
 
 | |
5AB6
 
 | |
5AB5
 
 | |
5AB4
 
 | |
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | 分子名称: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | 著者 | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
5A2O
 
 | |
5A2N
 
 | |
4ZVU
 
 | |
4ZVT
 
 | |
4ZVS
 
 | |
4ZVR
 
 | |
4ZVQ
 
 | |
4ZVP
 
 | |
4ZVO
 
 | |
4ZUZ
 
 | | SidC 1-871 | | 分子名称: | SidC | | 著者 | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | 登録日 | 2015-05-18 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
4ZGY
 
 | |
