8VAL
 
 | |
7AAD
 
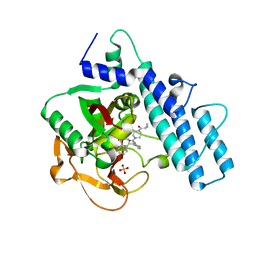 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | 分子名称: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
8WM7
 
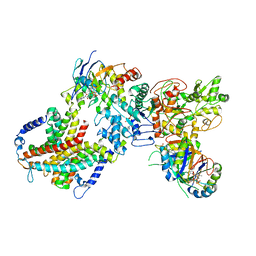 | | Cryo-EM structure of cyanobacterial nitrate/nitrite transporter NrtBCD in complex with signalling protein PII | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Nitrate transport ATP-binding protein, Nitrate transport permease protein, ... | | 著者 | Li, B, Zhou, C.Z, Chen, Y.X, Jiang, Y.L. | | 登録日 | 2023-10-03 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Allosteric regulation of nitrate transporter NRT via the signaling protein PII.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZOZ
 
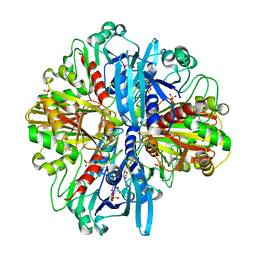 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | 著者 | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | 登録日 | 2024-05-29 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
6ZT8
 
 | | X-ray structure of mutated arabinofuranosidase | | 分子名称: | Alpha-L-arabinofuranosidase, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | 著者 | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | 登録日 | 2020-07-17 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
8VAQ
 
 | |
8VAP
 
 | |
8Y6V
 
 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | 分子名称: | gp119, gp120, gp162, ... | | 著者 | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | 登録日 | 2024-02-03 | | 公開日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
8VAT
 
 | |
8VZ9
 
 | | Crystal structure of mouse MAIT M2A TCR-MR1-5-OP-RU complex | | 分子名称: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Ciacchi, L, Rossjohn, J, Awad, W. | | 登録日 | 2024-02-11 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
4V2I
 
 | | Biochemical characterization and structural analysis of a new cold- active and salt tolerant esterase from the marine bacterium Thalassospira sp | | 分子名称: | ESTERASE/LIPASE, MAGNESIUM ION | | 著者 | Santi, C.D, Leiros, H.-K.S, Scala, A.D, Pascale, D.D, Altermark, B, Willassen, N.-P. | | 登録日 | 2014-10-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.686 Å) | | 主引用文献 | Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
Extremophiles, 20, 2016
|
|
8VAN
 
 | | Structure of the E. coli clamp loader bound to the beta clamp in an Initial-Binding conformation | | 分子名称: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | 著者 | Landeck, J.T, Pajak, J, Kelch, B.A. | | 登録日 | 2023-12-11 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication.
J.Biol.Chem., 300, 2024
|
|
9ER3
 
 | |
9C81
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | 分子名称: | (2R)-2-phenoxy-3-{[(1S,2S,4S)-spiro[bicyclo[2.2.1]heptane-7,1'-cyclopropane]-2-carbonyl]amino}propanoic acid, AmpC Beta-lactamase | | 著者 | Liu, F, Shoichet, B.K, Bassim, V. | | 登録日 | 2024-06-11 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
5JR8
 
 | | Disposal of Iron by a Mutant form of Siderocalin NGAL | | 分子名称: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, PHOSPHATE ION | | 著者 | Rupert, P.B, Strong, R.K, Barasch, J, Hollman, M, Deng, R, Hod, E.A, Abergel, R, Allred, B, Xu, K, Darrah, S, Tekabe, Y, Perlstein, A, Bruck, E, Stauber, J, Corbin, K, Buchen, C, Slavkovich, V, Graziano, J, Spitalnik, S, Qiu, A. | | 登録日 | 2016-05-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Disposal of iron by a mutant form of lipocalin 2.
Nat Commun, 7, 2016
|
|
9C6P
 
 | |
8WIU
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | 分子名称: | 7-[5-[1-(cyclopropylmethyl)-3,5-dimethyl-pyrazol-4-yl]pyridin-3-yl]-1~{H}-imidazo[4,5-b]pyridine, Isoform C of Bromodomain-containing protein 4 | | 著者 | Cao, D, Zhiyan, D, Xiong, B. | | 登録日 | 2023-09-25 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Discovery of a brain-permeable bromodomain and extra terminal domain (BET) inhibitor with selectivity for BD1 for the treatment of multiple sclerosis.
Eur.J.Med.Chem., 265, 2023
|
|
8VAM
 
 | |
9C8J
 
 | |
8VAS
 
 | |
9EM2
 
 | |
7P81
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
9FJK
 
 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | 分子名称: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | 著者 | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | 登録日 | 2024-05-31 | | 公開日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
8VAR
 
 | |
9FQ2
 
 | | Poliovirus 3C protease in H32 spacegroup | | 分子名称: | Protease 3C | | 著者 | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | 登録日 | 2024-06-14 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Poliovirus 3C protease in H32 spacegroup
To Be Published
|
|
