1GKM
 
 | |
5M2K
 
 | | Crystal structure of vancomycin-Zn(II) complex | | 分子名称: | 1,2-ETHANEDIOL, ZINC ION, vancomycin, ... | | 著者 | Zarkan, A, Macklyne, H.-R, Chirgadze, D.Y, Bond, A.D, Hesketh, A.R, Hong, H.-J. | | 登録日 | 2016-10-13 | | 公開日 | 2017-07-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Sci Rep, 7, 2017
|
|
2X46
 
 | | Crystal Structure of SeMet Arg r 1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALLERGEN ARG R 1 | | 著者 | Paesen, G.C, Siebold, C, Syme, N, Harlos, K, Graham, S.C, Hilger, C, Homans, S.W, Hentges, F, Stuart, D.I. | | 登録日 | 2010-01-28 | | 公開日 | 2011-02-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of the Allergen Arg R 1, a Histamine-Binding Lipocalin from a Soft Tick
To be Published
|
|
5IQN
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_SRIRIRGYVR | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, Protein FimF, ... | | 著者 | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | 登録日 | 2016-03-11 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4MTY
 
 | | Structure at 1A resolution of a helical aromatic foldamer-protein complex. | | 分子名称: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | 著者 | Ogayone, T, Buratto, J, Langlois D'Estaintot, B, Stupfel, M, Granier, T, Gallois, B, Huc, Y. | | 登録日 | 2013-09-20 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 a resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2IIM
 
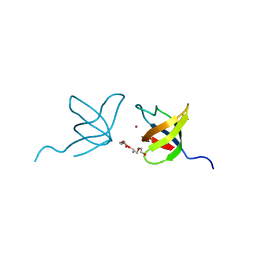 | | SH3 Domain of Human Lck | | 分子名称: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase LCK, TETRAETHYLENE GLYCOL, ... | | 著者 | Romir, J, Egerer-Sieber, C, Muller, Y.A. | | 登録日 | 2006-09-28 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal structure analysis and solution studies of human Lck-SH3; zinc-induced homodimerization competes with the binding of proline-rich motifs.
J.Mol.Biol., 365, 2007
|
|
8G22
 
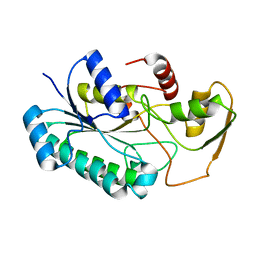 | | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae. | | 分子名称: | dTDP-4-dehydrorhamnose reductase | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2023-02-03 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae.
To Be Published
|
|
7TMJ
 
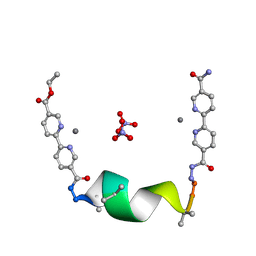 | |
7QUA
 
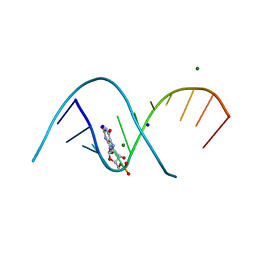 | | Duplex RNA containing Xanthosine-Cytosine base pairs | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*CP*GP*CP*GP*(XAN)P*AP*UP*UP*AP*GP*CP*G)-3'), SODIUM ION | | 著者 | Ennifar, E, Micura, R. | | 登録日 | 2022-01-17 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Towards a comprehensive understanding of RNA deamination: synthesis and properties of xanthosine-modified RNA.
Nucleic Acids Res., 50, 2022
|
|
1MSO
 
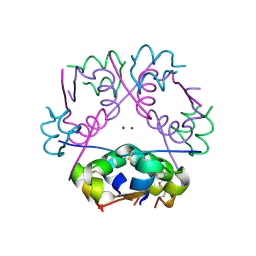 | | T6 Human Insulin at 1.0 A Resolution | | 分子名称: | Insulin A-Chain, Insulin B-Chain, ZINC ION | | 著者 | Smith, G.D, Pangborn, W.A, Blessing, R.H. | | 登録日 | 2002-09-19 | | 公開日 | 2003-03-04 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | The structure of T6 human insulin at 1.0 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5SV5
 
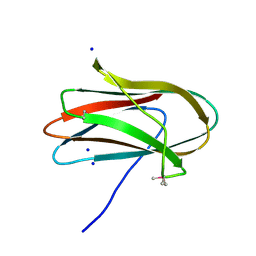 | | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis. | | 分子名称: | Microbial collagenase, SODIUM ION | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-08-04 | | 公開日 | 2016-08-17 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis.
To Be Published
|
|
2JHF
 
 | | Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase | | 分子名称: | ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | 著者 | Meijers, R, Adolph, H.W, Dauter, Z, Wilson, K.S, Lamzin, V.S, Cedergren-Zeppezauer, E.S. | | 登録日 | 2007-02-22 | | 公開日 | 2007-04-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structural Evidence for a Ligand Coordination Switch in Liver Alcohol Dehydrogenase
Biochemistry, 46, 2007
|
|
1N4V
 
 | |
1C0R
 
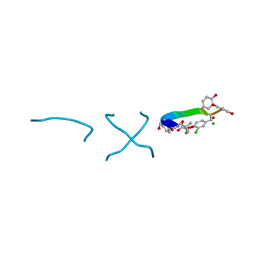 | | COMPLEX OF VANCOMYCIN WITH D-LACTIC ACID | | 分子名称: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | 著者 | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | 登録日 | 1999-07-20 | | 公開日 | 1999-07-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Vancomycin Binding to Low-Affinity Ligands: Delineating a Minimum Set of Interactions Necessary for High-Affinity Binding.
J.Med.Chem., 42, 1999
|
|
1UZV
 
 | | High affinity fucose binding of Pseudomonas aeruginosa lectin II: 1.0 A crystal structure of the complex | | 分子名称: | CALCIUM ION, PSEUDOMONAS AERUGINOSA LECTIN II, SULFATE ION, ... | | 著者 | Mitchell, E, Sabin, C.D, Snajdrova, L, Budova, M, Perret, S, Gautier, C, Gilboa-Garber, N, Koca, J, Wimmerova, M, Imberty, A. | | 登録日 | 2004-03-17 | | 公開日 | 2004-12-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | High Affinity Fucose Binding of Pseudomonas Aeruginosa Lectin Pa-Iil: 1.0 A Resolution Crystal Structure of the Complex Combined with Thermodynamics and Computational Chemistry Approaches.
Proteins, 58, 2005
|
|
1YRI
 
 | | Chicken villin subdomain HP-35, N68H, pH6.4 | | 分子名称: | ACETATE ION, IODIDE ION, Villin | | 著者 | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | 登録日 | 2005-02-03 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6I7Y
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT56 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [1-[4-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]phenyl]piperidin-4-yl]-trimethyl-azanium | | 著者 | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of the first bromodomain of BRD4 in complex with RT56
To Be Published
|
|
4YNH
 
 | |
3P8J
 
 | |
5MAJ
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[cyclopentyl(imidazo[1,2-a]pyridin-2-ylmethyl)amino]-6-morpholino-1,3,5-triazine-2-carbonitrile | | 分子名称: | 1,2-ETHANEDIOL, Cathepsin L1, ~{N}-cyclopentyl-~{N}-(imidazo[1,2-a]pyridin-2-ylmethyl)-4-(iminomethyl)-6-morpholin-4-yl-1,3,5-triazin-2-amine | | 著者 | Kuglstatter, A, Stihle, M, Benz, J. | | 登録日 | 2016-11-03 | | 公開日 | 2017-01-11 | | 最終更新日 | 2017-02-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
2CNQ
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | 著者 | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | 登録日 | 2006-05-23 | | 公開日 | 2006-06-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
1YWC
 
 | | Structure of the ferrous CO complex of NP4 from Rhodnius Prolixus at pH 7.0 | | 分子名称: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, nitrophorin 4 | | 著者 | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | 登録日 | 2005-02-17 | | 公開日 | 2005-10-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
5D66
 
 | |
2VHA
 
 | | DEBP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, PERIPLASMIC BINDING TRANSPORT PROTEIN | | 著者 | Hu, Y.L, Fan, C.-P, Fu, G.S, Zhu, D.Y, Jin, Q, Wang, D.-C. | | 登録日 | 2007-11-20 | | 公開日 | 2008-07-08 | | 最終更新日 | 2019-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution.
J.Mol.Biol., 382, 2008
|
|
3JYO
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Niefind, K, Schomburg, D. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
