3D18
 
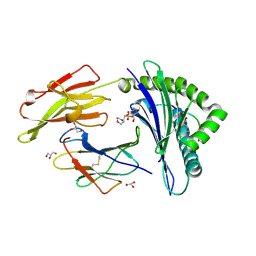 | | Crystal structure of HLA-B*2709 complexed with a variant of the latent membrane protein 2 peptide (LMP2(L)) of epstein-barr virus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | 著者 | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | 登録日 | 2008-05-05 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal structure of HLA-B*2709 complexed with a variant of the latent membrane protein 2 peptide (LMP2(L)) of epstein-barr virus
To be Published
|
|
3D4Z
 
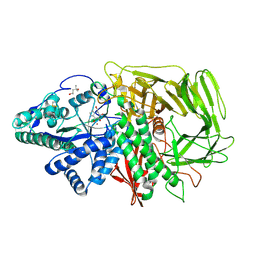 | | GOLGI MANNOSIDASE II complex with gluco-imidazole | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, GLUCOIMIDAZOLE, ... | | 著者 | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
5RPW
 
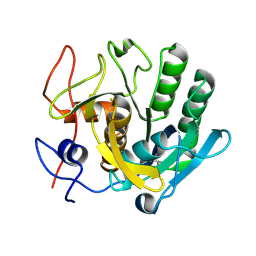 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo63 | | 分子名称: | Proteinase K | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-23 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3D3F
 
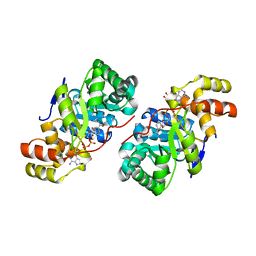 | |
3D3T
 
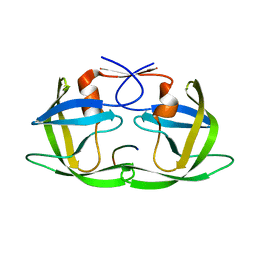 | | Crystal Structure of HIV-1 CRF01_AE in complex with the substrate p1-p6 | | 分子名称: | HIV-1 protease, P1-P6 SUBSTRATE PEPTIDE | | 著者 | Bandaranayake, R.M, Prabu-Jeyabalan, M, Kakizawa, J, Sugiura, W, Schiffer, C.A. | | 登録日 | 2008-05-12 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6.
J.Virol., 82, 2008
|
|
3CH3
 
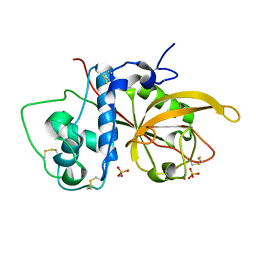 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum | | 分子名称: | DIHYDROGENPHOSPHATE ION, POTASSIUM ION, Serine-repeat antigen protein | | 著者 | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | 登録日 | 2008-03-07 | | 公開日 | 2008-03-25 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Insights into the Protease-like Antigen Plasmodium falciparum SERA5 and Its Noncanonical Active-Site Serine
J.Mol.Biol., 392, 2009
|
|
3CK2
 
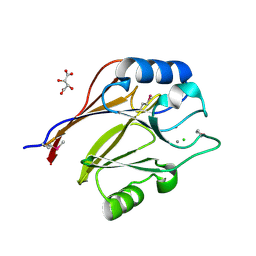 | | Crystal structure of conserved uncharacterized protein (predicted phosphoesterase COG0622) from Streptococcus pneumoniae TIGR4 | | 分子名称: | CHLORIDE ION, Conserved uncharacterized protein (predicted phosphoesterase COG0622), MANGANESE (II) ION, ... | | 著者 | Nocek, B, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-01 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of conserved uncharacterized protein (predicted phosphoesterase COG0622) from Streptococcus pneumoniae TIGR4.
To be Published
|
|
3CMG
 
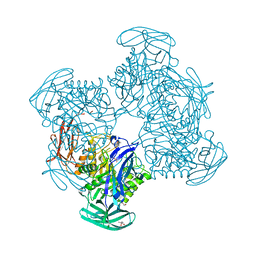 | | Crystal structure of putative beta-galactosidase from Bacteroides fragilis | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | Ramagopal, U.A, Rutter, M, Toro, R, Hu, S, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-03-21 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of putative beta-galactosidase from Bacteroides fragilis.
To be published
|
|
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | 分子名称: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | 著者 | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-03-27 | | 公開日 | 2008-04-08 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
5RSF
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000026180281 | | 分子名称: | 9-methyl-9H-purine-2,6-diamine, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSX
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388262 | | 分子名称: | 2-(3-methoxy-4-oxidanyl-phenyl)ethanoic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000157108 | | 分子名称: | 3-HYDROXYPICOLINIC ACID, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTT
 
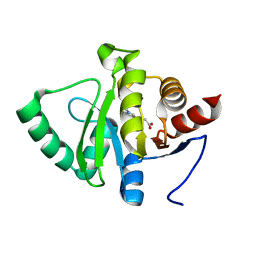 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000873830 | | 分子名称: | 3-(1-methyl-1H-indol-3-yl)propanoic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3CVT
 
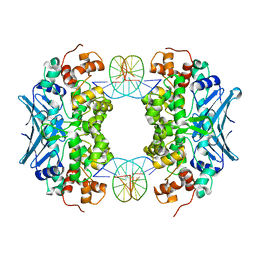 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | 分子名称: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*(8OG)P*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | 著者 | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | 登録日 | 2008-04-19 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
5RU9
 
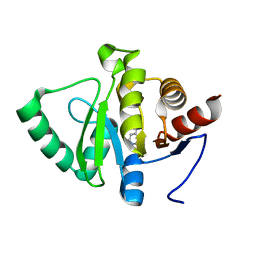 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000165882 | | 分子名称: | 3-AMINOPYRIDINE-4-CARBOXYLIC ACID, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUN
 
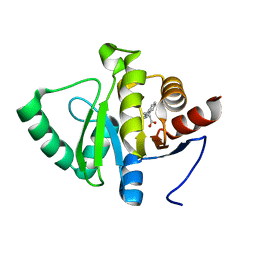 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000194295 | | 分子名称: | 3-(1H-benzimidazol-1-yl)propanoic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV5
 
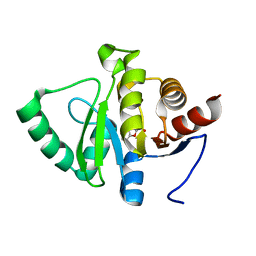 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008578948 | | 分子名称: | BENZOFURO[3,2-D]PYRIMIDIN-4(3H)-ONE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSN
 
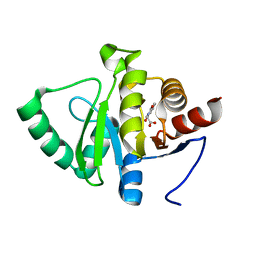 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000064576 | | 分子名称: | (1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3CTD
 
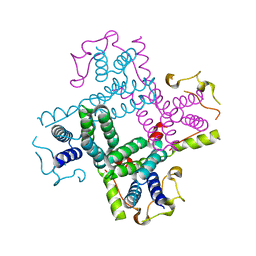 | | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris | | 分子名称: | Putative ATPase, AAA family | | 著者 | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-11 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris.
To be Published
|
|
5RVI
 
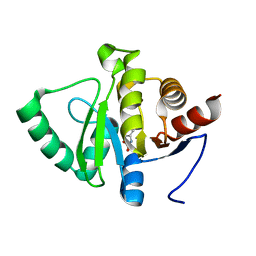 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000084843283 | | 分子名称: | CHLORZOXAZONE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (0.94 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RT3
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000039281982 | | 分子名称: | 1-benzothiophen-2-ylacetic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTK
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000164504 | | 分子名称: | 1,3-benzodioxol-5-ol, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU1
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000034687 | | 分子名称: | DIPHENYLACETIC ACID, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3D0E
 
 | | Crystal structure of human Akt2 in complex with GSK690693 | | 分子名称: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3-yn-2-ol, RAC-beta serine/threonine-protein kinase | | 著者 | Concha, N.O, Smallwood, A. | | 登録日 | 2008-05-01 | | 公開日 | 2008-10-21 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H-imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase.
J.Med.Chem., 51, 2008
|
|
5RUI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000332651 | | 分子名称: | Non-structural protein 3, isoquinolin-1(2H)-one | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
