1N17
 
 | | Structure and Dynamics of Thioguanine-modified Duplex DNA | | 分子名称: | 5'-D(*GP*CP*TP*AP*AP*GP*(S6G)P*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | 著者 | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | 登録日 | 2002-10-16 | | 公開日 | 2002-10-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
2PJI
 
 | |
1Y9H
 
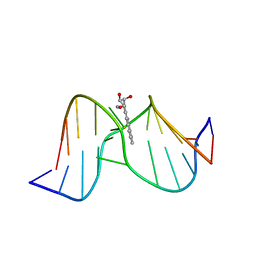 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | 分子名称: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | 著者 | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 2004-12-15 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-04-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
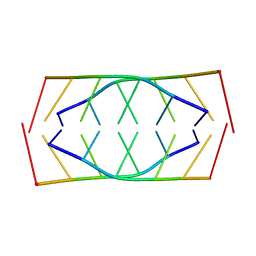
1YBR
 
 | |
2AIT
 
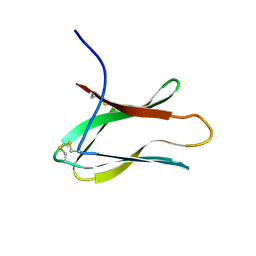 | | DETERMINATION OF THE COMPLETE THREE-DIMENSIONAL STRUCTURE OF THE ALPHA-AMYLASE INHIBITOR TENDAMISTAT IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE AND DISTANCE GEOMETRY | | 分子名称: | TENDAMISTAT | | 著者 | Kline, A.D, Braun, W, Guntert, P, Billeter, M, Wuthrich, K. | | 登録日 | 1989-05-24 | | 公開日 | 1990-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Determination of the complete three-dimensional structure of the alpha-amylase inhibitor tendamistat in aqueous solution by nuclear magnetic resonance and distance geometry.
J.Mol.Biol., 204, 1988
|
|
1MVK
 
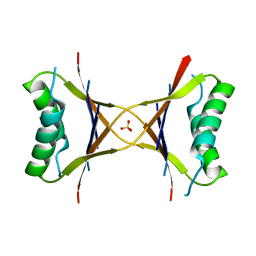 | | X-ray structure of the tetrameric mutant of the B1 domain of streptococcal protein G | | 分子名称: | Immunoglobulin G binding protein G, SULFATE ION | | 著者 | Frank, M.K, Dyda, F, Dobrodumov, A, Gronenborn, A.M. | | 登録日 | 2002-09-25 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Core mutations switch monomeric protein GB1 into an intertwined tetramer.
Nat.Struct.Biol., 9, 2002
|
|
1MYQ
 
 | | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction | | 分子名称: | 5'-D(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3' | | 著者 | Matsugami, A, Ouhashi, K, Kanagawa, M, Liu, H, Kanagawa, S, Uesugi, S, Katahira, M. | | 登録日 | 2002-10-04 | | 公開日 | 2002-10-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction.
J.Mol.Biol., 313, 2001
|
|
1YE3
 
 | |
1YBN
 
 | |
1KPY
 
 | |
2O49
 
 | |
1KPZ
 
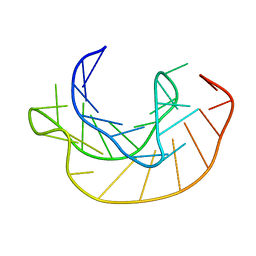 | |
2AWQ
 
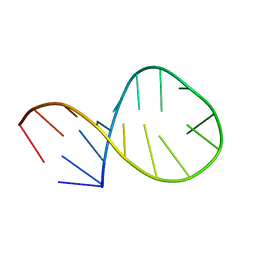 | |
1MSJ
 
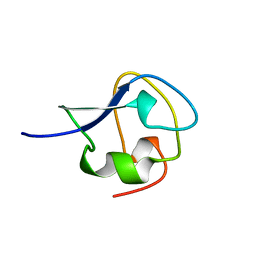 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | 分子名称: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | 著者 | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | 登録日 | 1999-01-24 | | 公開日 | 1999-04-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1N14
 
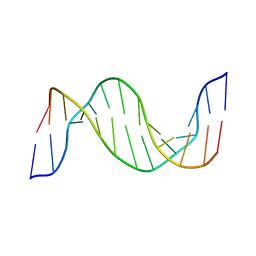 | | Structure and Dynamics of Thioguanine-modified Duplex DNA in Comparison with Unmodified DNA; Structure of Unmodified Duplex DNA | | 分子名称: | 5'-D(*GP*CP*TP*AP*AP*GP*GP*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | 著者 | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | 登録日 | 2002-10-16 | | 公開日 | 2002-10-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
1YSX
 
 | | Solution structure of domain 3 from human serum albumin complexed to an anti-apoptotic ligand directed against Bcl-xL and Bcl-2 | | 分子名称: | 4-({2-[(2,4-DIMETHYLPHENYL)SULFANYL]ETHYL}AMINO)-N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITROBENZENESULFONAMIDE, Serum albumin | | 著者 | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | 登録日 | 2005-02-09 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSI
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | 分子名称: | Apoptosis regulator Bcl-X, N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITRO-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE | | 著者 | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | 登録日 | 2005-02-08 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1LVR
 
 | | IC3 of CB1 (L431A,A432L) Bound to G(alpha)i | | 分子名称: | Cannabinoid receptor 1 | | 著者 | Ulfers, A.L, McMurry, J.L, Miller, A, Wang, L, Kendall, D.A, Mierke, D.F. | | 登録日 | 2002-05-29 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cannabinoid receptor-G protein interactions: G(alphai1)-bound structures of IC3 and a mutant with altered G protein specificity.
Protein Sci., 11, 2002
|
|
1YWX
 
 | | Solution Structure of Methanococcus maripaludis Protein MMP0443: The Northeast Structural Genomics Consortium Target MrR16 | | 分子名称: | 30S ribosomal protein S24e | | 著者 | Yang, C.S, Acton, T, Shen, Y, Ma, L, Liu, G, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-02-18 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Methanococcus maripaludis Protein MMP0443: The Northeast Structural Genomics Consortium Target MrR16
To be Published
|
|
1KBR
 
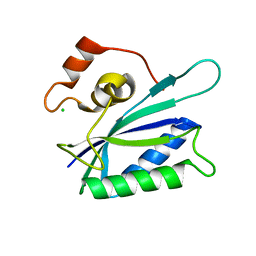 | |
1X6M
 
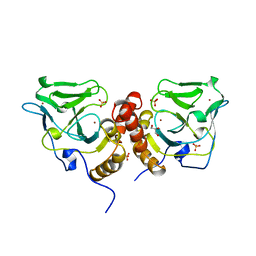 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | 分子名称: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | 著者 | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | 登録日 | 2004-08-11 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1XA8
 
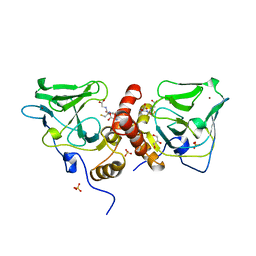 | | Crystal Structure Analysis of Glutathione-dependent formaldehyde-activating enzyme (Gfa) | | 分子名称: | GLUTATHIONE, GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, ... | | 著者 | Neculai, A.M, Neculai, D, Griesinger, C, Vorholt, J.A, Becker, S. | | 登録日 | 2004-08-25 | | 公開日 | 2004-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1N51
 
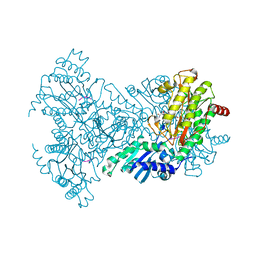 | | Aminopeptidase P in complex with the inhibitor apstatin | | 分子名称: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | 著者 | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | 登録日 | 2002-11-03 | | 公開日 | 2003-12-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NJ3
 
 | | Structure and Ubiquitin Interactions of the Conserved NZF Domain of Npl4 | | 分子名称: | NPL4, ZINC ION | | 著者 | Wang, B, Alam, S.L, Meyer, H.H, Payne, M, Stemmler, T.L, Davis, D.R, Sundquist, W.I. | | 登録日 | 2002-12-30 | | 公開日 | 2003-04-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.
J.Biol.Chem., 278, 2003
|
|
2AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14Q | | 分子名称: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | 著者 | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | 登録日 | 1999-01-18 | | 公開日 | 1999-04-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
