6QAM
 
 | |
6QEB
 
 | |
2ZNF
 
 | | HIGH-RESOLUTION STRUCTURE OF AN HIV ZINC FINGERLIKE DOMAIN VIA A NEW NMR-BASED DISTANCE GEOMETRY APPROACH | | 分子名称: | GAG POLYPROTEIN, ZINC ION | | 著者 | Summers, M.F, South, T.L, Kim, B, Hare, D.R. | | 登録日 | 1990-03-30 | | 公開日 | 1991-10-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach.
Biochemistry, 29, 1990
|
|
6C00
 
 | |
2XKM
 
 | | Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR | | 分子名称: | CAPSID PROTEIN G8P | | 著者 | Straus, S.K, P Scott, W.R, Schwieters, C.D, Marvin, D.A. | | 登録日 | 2010-07-09 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | FIBER DIFFRACTION (3.3 Å), SOLID-STATE NMR | | 主引用文献 | Consensus Structure of Pf1 Filamentous Bacteriophage from X-Ray Fibre Diffraction and Solid-State NMR.
Eur.Biophys.J., 40, 2011
|
|
6GZK
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | 分子名称: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | 著者 | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | 登録日 | 2018-07-04 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
7LOH
 
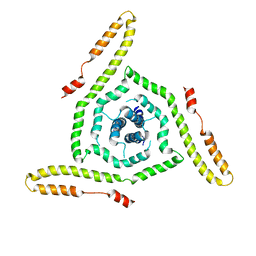 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail | | 分子名称: | Transmembrane protein gp41 | | 著者 | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | 登録日 | 2021-02-10 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | 分子名称: | ATP synthase subunit c | | 著者 | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | 登録日 | 2021-12-23 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
6TTA
 
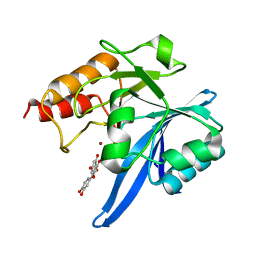 | | Haddock model of NDM-1/quercetin complex | | 分子名称: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Metallo beta lactamase NDM-1, ZINC ION | | 著者 | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | 登録日 | 2019-12-26 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6X8R
 
 | |
6YJ0
 
 | |
7LOI
 
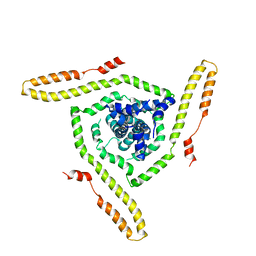 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail | | 分子名称: | Transmembrane protein gp41 | | 著者 | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | 登録日 | 2021-02-10 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
1MPH
 
 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | 分子名称: | BETA SPECTRIN | | 著者 | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | 登録日 | 1997-04-23 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
1BQT
 
 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | 分子名称: | INSULIN-LIKE GROWTH FACTOR-I | | 著者 | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | 登録日 | 1998-08-18 | | 公開日 | 1999-05-18 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
6TTC
 
 | | Haddock model of NDM-1/myricetin complex | | 分子名称: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | 著者 | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | 登録日 | 2019-12-26 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TT8
 
 | | Haddock model of NDM-1/morin complex | | 分子名称: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | 著者 | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | 登録日 | 2019-12-25 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6U6P
 
 | |
6U6R
 
 | |
6U6S
 
 | |
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZC
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZB
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
6GZR
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | 分子名称: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | 著者 | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | 登録日 | 2018-07-05 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
1AGO
 
 | |
1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | 分子名称: | TRYPSIN INHIBITOR V | | 著者 | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | 登録日 | 1995-10-26 | | 公開日 | 1996-04-03 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
