4V5J
 
 | | Structure of the 70S ribosome bound to Release factor 2 and a substrate analog provides insights into catalysis of peptide release | | 分子名称: | 16S Ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Jin, H, Kelley, A.C, Loakes, D, Ramakrishnan, V. | | 登録日 | 2010-03-24 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
5SOV
 
 | |
4V7Y
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with azithromycin. | | 分子名称: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | 登録日 | 2010-08-18 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2STB
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | 分子名称: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | 著者 | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | 登録日 | 1998-12-11 | | 公開日 | 2000-01-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5SRA
 
 | |
8H59
 
 | | A fungal MAP kinase in complex with an inhibitor | | 分子名称: | Mitogen-activated protein kinase MPS1, ~{N}-[(2~{S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]-8-[2-methoxy-5-(trifluoromethyloxy)phenyl]-1,6-naphthyridine-2-carboxamide | | 著者 | Kong, Z, Zhang, X, Wang, D, Liu, J. | | 登録日 | 2022-10-12 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi.
Mbio, 14, 2023
|
|
5SR9
 
 | |
2THF
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225F MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | 分子名称: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, THROMBIN HEAVY CHAIN, ... | | 著者 | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | 登録日 | 1999-01-26 | | 公開日 | 1999-03-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8H9B
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 3 | | 分子名称: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | 著者 | Qiao, H, Xia, M, Wang, J, Fang, P. | | 登録日 | 2022-10-25 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
4V9F
 
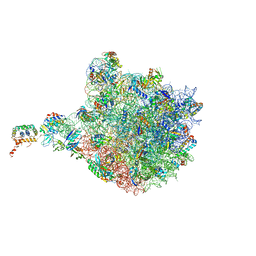 | |
5SRC
 
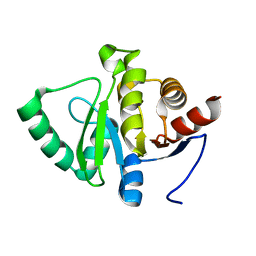 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5198562500 - (R,R) and (R,S) isomers | | 分子名称: | (2R)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid, (2S)-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)piperidin-3-yl](hydroxy)acetic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2RC9
 
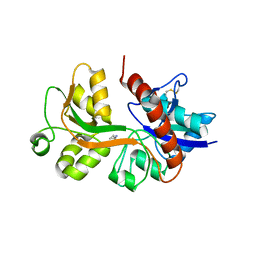 | |
8HHZ
 
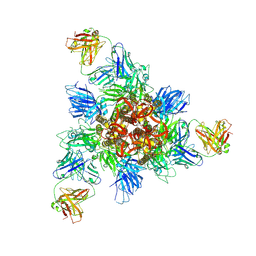 | |
4WDG
 
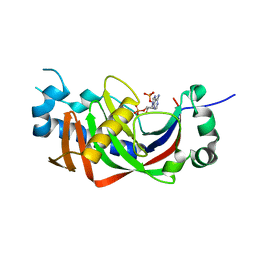 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, complexed with 2',5'-ADP | | 分子名称: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-5'-DIPHOSPHATE | | 著者 | Myllykoski, M, Raasakka, A, Kursula, P. | | 登録日 | 2014-09-08 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
5SON
 
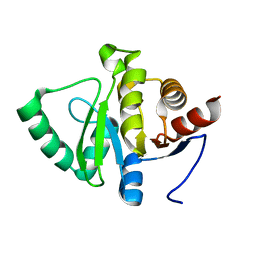 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000920153280 - (R) isomer | | 分子名称: | 3-{[(2R)-2-phenylpropyl]sulfanyl}-7H-[1,2,4]triazolo[4,3-b][1,2,4]triazole, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H8J
 
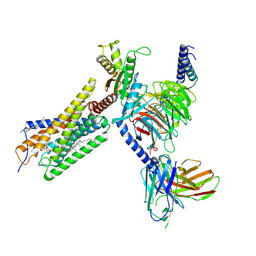 | | Lodoxamide-bound GPR35 in complex with G13 | | 分子名称: | 2,2'-[(2-chloro-5-cyano-1,3-phenylene)bis(azanediyl)]bis(oxoacetic acid), CALCIUM ION, CHOLESTEROL, ... | | 著者 | Yuan, Q, Duan, J, Liu, Q, Xu, H.E, Jiang, Y. | | 登録日 | 2022-10-23 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Insights into divalent cation regulation and G 13 -coupling of orphan receptor GPR35.
Cell Discov, 8, 2022
|
|
8HHO
 
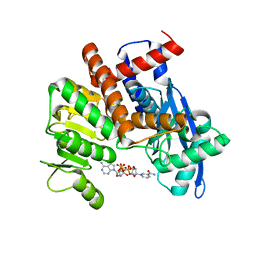 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
4WE0
 
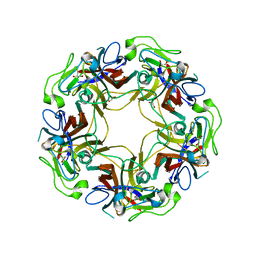 | | JC Polyomavirus VP1 five-fold pore mutant P223M | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | 著者 | Stroh, L.J, Stehle, T. | | 登録日 | 2014-09-09 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Modulation of a Pore in the Capsid of JC Polyomavirus Reduces Infectivity and Prevents Exposure of the Minor Capsid Proteins.
J.Virol., 89, 2015
|
|
5SOS
 
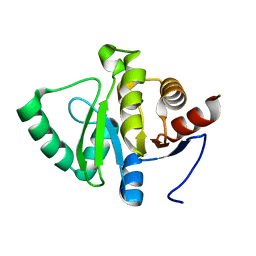 | |
4UQI
 
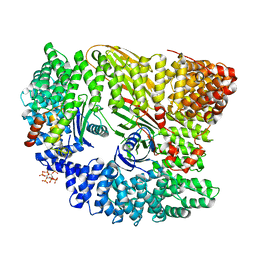 | | AP2 controls clathrin polymerization with a membrane-activated switch | | 分子名称: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA, AP-2 COMPLEX SUBUNIT MU, ... | | 著者 | Kelly, B.T, Graham, S.C, Liska, N, Dannhauser, P.N, Hoening, S, Ungewickell, E.J, Owen, D.J. | | 登録日 | 2014-06-23 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Clathrin Adaptors. Ap2 Controls Clathrin Polymerization with a Membrane-Activated Switch.
Science, 345, 2014
|
|
2TMV
 
 | |
5SOW
 
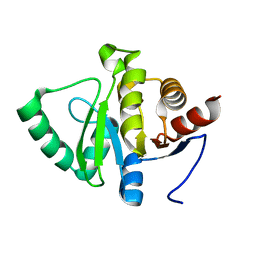 | |
8HIQ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-21 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
5SRD
 
 | |
2TPK
 
 | |
