6W7K
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ634p | | 分子名称: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyridin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | 著者 | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
8P9O
 
 | |
5Z00
 
 | | AtVAL1 B3 domain in complex with 15bp-DNA | | 分子名称: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), ... | | 著者 | Wu, B.X, Zhang, M.M. | | 登録日 | 2017-12-17 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.587 Å) | | 主引用文献 | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
6W7T
 
 | | Structure of PaP3 small terminase | | 分子名称: | small terminase subunit | | 著者 | Cingolani, G, Lokareddy, R. | | 登録日 | 2020-03-19 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
6TNG
 
 | |
6TNI
 
 | |
1F6O
 
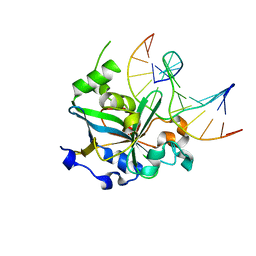 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH DNA | | 分子名称: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(YRR)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | 著者 | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | 登録日 | 2000-06-22 | | 公開日 | 2000-12-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5ZCW
 
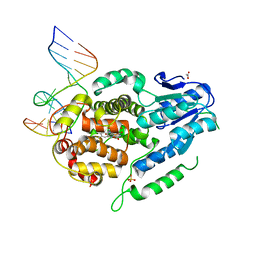 | | Structure of the Methanosarcina mazei class II CPD-photolyase in complex with intact, phosphodiester linked, CPD-lesion | | 分子名称: | 5'-D(*AP*TP*CP*GP*GP*CP*(TTD)P*CP*GP*CP*GP*CP*AP*A)-3', 5'-D(*TP*GP*CP*GP*CP*GP*AP*AP*GP*CP*CP*GP*AP*T)-3', ACETATE ION, ... | | 著者 | Maestre-Reyna, M, Bessho, Y. | | 登録日 | 2018-02-21 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.
IUCrJ, 5, 2018
|
|
7E1P
 
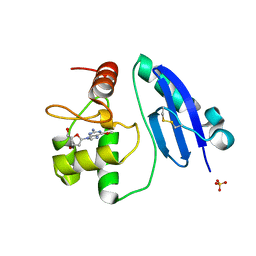 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase C120S variant in complex with O6-methyldeoxyguanosine | | 分子名称: | (2~{R},3~{S},5~{R})-5-(2-azanyl-6-methoxy-purin-9-yl)-2-(hydroxymethyl)oxolan-3-ol, Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | 著者 | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | 登録日 | 2021-02-03 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7DUV
 
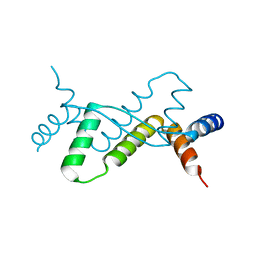 | | Structure of Sulfolobus solfataricus SegB protein | | 分子名称: | SULFATE ION, SegB | | 著者 | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | 登録日 | 2021-01-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7C7Y
 
 | | C-terminal domain of B. cereus TubY | | 分子名称: | Uncharacterized protein | | 著者 | Hayashi, I. | | 登録日 | 2020-05-27 | | 公開日 | 2020-10-28 | | 最終更新日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The C-terminal region of the plasmid partitioning protein TubY is a tetramer that can bind membranes and DNA.
J.Biol.Chem., 295, 2020
|
|
1D8Y
 
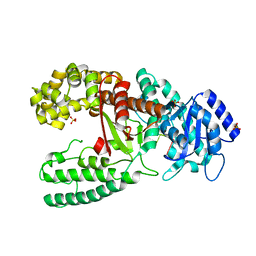 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA | | 分子名称: | D(T)19 OLIGOMER, DNA POLYMERASE I, SULFATE ION, ... | | 著者 | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | 登録日 | 1999-10-26 | | 公開日 | 1999-12-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7DQQ
 
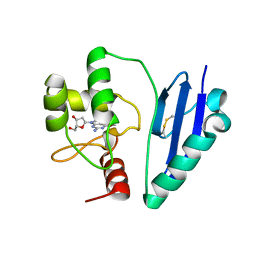 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase Y91F/C120S variant in complex with O6-methyldeoxyguanosine | | 分子名称: | (2~{R},3~{S},5~{R})-5-(2-azanyl-6-methoxy-purin-9-yl)-2-(hydroxymethyl)oxolan-3-ol, Methylated-DNA--protein-cysteine methyltransferase | | 著者 | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | 登録日 | 2020-12-24 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1D9D
 
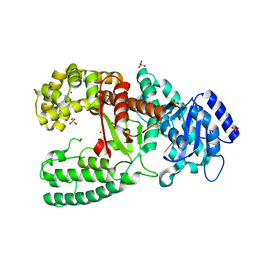 | | CRYSTALL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH SHORT DNA FRAGMENT CARRYING 2'-0-AMINOPROPYL-RNA MODIFICATIONS 5'-D(TCG)-AP(AUC)-3' | | 分子名称: | 5'-D(*TP*CP*GP)-R(AP*(U31)P*(C31))-3', DNA POLYMERASE I, MAGNESIUM ION, ... | | 著者 | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | 登録日 | 1999-10-27 | | 公開日 | 1999-12-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4JS4
 
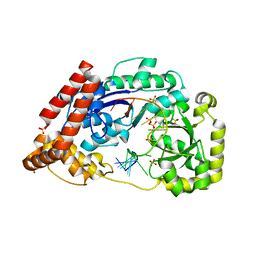 | |
4JS5
 
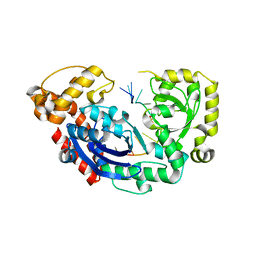 | |
8RPX
 
 | | NhoI restriction endonuclease in complex with quadruply methylated DNA target | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*TP*GP*(5CM)P*AP*GP*(5CM)P*TP*C)-3'), ... | | 著者 | Rafalski, D, Krakowska, K, Gilski, M, Bochtler, M. | | 登録日 | 2024-01-17 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural analysis of the BisI family of modification dependent restriction endonucleases.
Nucleic Acids Res., 52, 2024
|
|
8T5Y
 
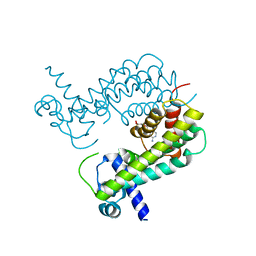 | |
8HCC
 
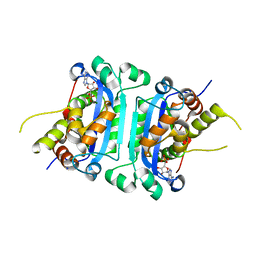 | | Crystal structure of mTREX1-RNA product complex (AMP) | | 分子名称: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | 登録日 | 2022-11-01 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
1EXQ
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | 分子名称: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | 著者 | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | 登録日 | 2000-05-03 | | 公開日 | 2000-11-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3VGZ
 
 | | Crystal structure of E. coli YncE | | 分子名称: | Uncharacterized protein YncE | | 著者 | Kagawa, W, Sagawa, T, Niki, H, Kurumizaka, H. | | 登録日 | 2011-08-23 | | 公開日 | 2011-11-16 | | 最終更新日 | 2011-12-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE
Acta Crystallogr.,Sect.D, 67, 2011
|
|
8IIO
 
 | | H109Q mutant of uracil DNA glycosylase X | | 分子名称: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIM
 
 | | H109K mutant of uracil DNA glycosylase X | | 分子名称: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIT
 
 | |
8IIH
 
 | | H109C mutant of uracil DNA glycosylase X | | 分子名称: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
