7LEY
 
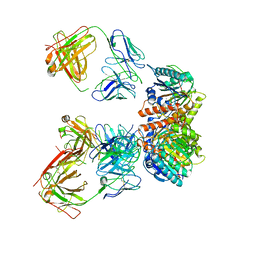 | | Trimeric human Arginase 1 in complex with mAb5 | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb5 heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEX
 
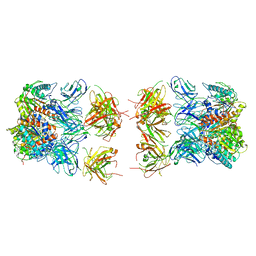 | |
6Q37
 
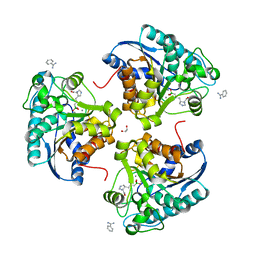 | | Complex of Arginase 2 with Example 23 | | 分子名称: | 1,2-ETHANEDIOL, 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, ... | | 著者 | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | 登録日 | 2018-12-03 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.211 Å) | | 主引用文献 | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6Q3F
 
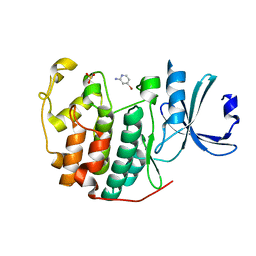 | | CDK2 in complex with FragLite2 | | 分子名称: | 4-bromanylpyridin-2-amine, Cyclin-dependent kinase 2 | | 著者 | Wood, D.J, Martin, M.P, Noble, M.E.M. | | 登録日 | 2018-12-04 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7LPO
 
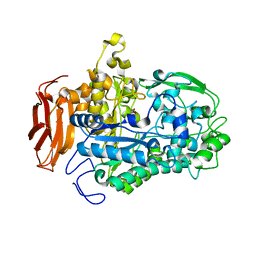 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with tris | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytoplasmic protein, MAGNESIUM ION | | 著者 | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | 登録日 | 2021-02-12 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LF0
 
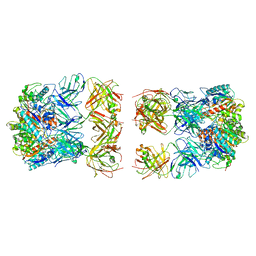 | |
7LPP
 
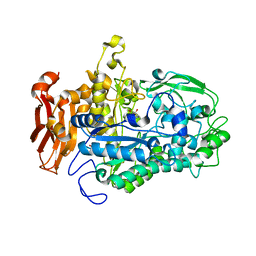 | | Crystal structure of Cryptococcus neoformans sterylglucosidase 1 with hit 1 | | 分子名称: | 4-(hydroxymethyl)-1-[[2-(3-methoxyphenyl)-1,3-thiazol-5-yl]methyl]piperidin-4-ol, Cytoplasmic protein, GLYCEROL, ... | | 著者 | Pereira de Sa, N, Del Poeta, M, Airola, M.V. | | 登録日 | 2021-02-12 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure and inhibition of Cryptococcus neoformans sterylglucosidase to develop antifungal agents.
Nat Commun, 12, 2021
|
|
7LF2
 
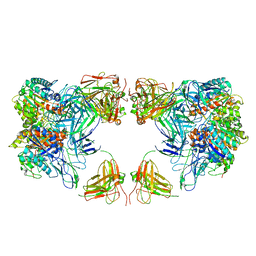 | | Trimeric human Arginase 1 in complex with mAb4 | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb4 monoclonal antibody heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
1ML3
 
 | | Evidences for a flip-flop catalytic mechanism of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase, from its crystal structure in complex with reacted irreversible inhibitor 2-(2-phosphono-ethyl)-acrylic acid 4-nitro-phenyl ester | | 分子名称: | (3-FORMYL-BUT-3-ENYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | 著者 | Castilho, M.S, Pavao, F, Oliva, G. | | 登録日 | 2002-08-29 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evidence for the Two Phosphate Binding Sites of an Analogue of the Thioacyl Intermediate for the Trypanosoma cruzi Glyceraldehyde-3-phosphate Dehydrogenase-Catalyzed Reaction, from Its Crystal Structure.
Biochemistry, 42, 2003
|
|
4O5B
 
 | |
7LF1
 
 | | Trimeric human Arginase 1 in complex with mAb3 | | 分子名称: | Arginase-1, MANGANESE (II) ION, mAb3 heavy chain, ... | | 著者 | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | 登録日 | 2021-01-15 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
4O5H
 
 | |
4O1Z
 
 | | Crystal Structure of Ovine Cyclooxygenase-1 Complex with Meloxicam | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,3-thiazol-2-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | 著者 | Xu, S, Hermanson, D.J, Banerjee, S, Ghebreselasie, K, Clayton, G.M, Garavito, R.M, Marnett, L.J. | | 登録日 | 2013-12-16 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
7M1R
 
 | |
3UFC
 
 | |
4O5M
 
 | |
1MFU
 
 | | Probing the role of a mobile loop in human salivary amylase: Structural studies on the loop-deleted mutant | | 分子名称: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | 著者 | Ramasubbu, N, Ragunath, C, Mishra, P.J. | | 登録日 | 2002-08-13 | | 公開日 | 2002-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Probing the role of a mobile loop in substrate binding and enzyme activity of
human salivary amylase.
J.Mol.Biol., 325, 2003
|
|
6QGV
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with a Spiro[4.5]decanone inhibitor (JPHM-2-167) | | 分子名称: | 8-[(3-methylpyridin-2-yl)methyl]-3-(4-phenylphenyl)-1-pyrimidin-2-yl-1,3,8-triazaspiro[4.5]decane-2,4-dione, Egl nine homolog 1, GLYCEROL, ... | | 著者 | Chowdhury, R, Holt-Martyn, J.P, Rahman, M.Z, Schofield, C.J. | | 登録日 | 2019-01-13 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Studies on spiro[4.5]decanone prolyl hydroxylase domain inhibitors.
Medchemcomm, 10, 2019
|
|
4O34
 
 | | Room temperature macromolecular serial crystallography using synchrotron radiation | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Stellato, F, Oberthuer, D, Liang, M, Bean, R, Gati, C, Yefanov, O, Barty, A, Burkhardt, A, Fischer, P, Galli, L, Kirian, R.A, Mayer, J, Pannerselvam, S, Yoon, C.H, Chervinskii, F, Speller, E, White, T.A, Betzel, C, Meents, A, Chapman, H.N. | | 登録日 | 2013-12-18 | | 公開日 | 2014-06-11 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Room-temperature macromolecular serial crystallography using synchrotron radiation.
IUCrJ, 1, 2014
|
|
3ULL
 
 | |
7QHD
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | 分子名称: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | 登録日 | 2021-12-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QHE
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | 分子名称: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | 登録日 | 2021-12-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
4O46
 
 | | 14-3-3-gamma in complex with influenza NS1 C-terminal tail phosphorylated at S228 | | 分子名称: | 14-3-3 protein gamma, Nonstructural protein 1, UNKNOWN ATOM OR ION, ... | | 著者 | Qin, S, Liu, Y, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-18 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
1MI3
 
 | | 1.8 Angstrom structure of xylose reductase from Candida tenuis in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | 著者 | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | 登録日 | 2002-08-21 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases
Biochem.J., 373, 2003
|
|
7QNM
 
 | | Crystallization and structural analyses of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans | | 分子名称: | (S)-2-haloacid dehalogenase, PHOSPHATE ION | | 著者 | Grigorian, E, Roret, T, Leblanc, C, Delage, L, Czjzek, M. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | X-ray structure and mechanism of ZgHAD, a l-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 32, 2023
|
|
