5V6G
 
 | | Crystal structure of Influenza A virus Matrix Protein M1(NLS-88R) | | 分子名称: | Matrix protein 1 | | 著者 | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | 登録日 | 2017-03-16 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
5V8A
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88R, pH 7.3) | | 分子名称: | Matrix protein 1 | | 著者 | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Zhou, Q, Chiang, M.-J, Kosikova, M. | | 登録日 | 2017-03-21 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
5V7B
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E) | | 分子名称: | Matrix protein 1 | | 著者 | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | 登録日 | 2017-03-20 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
5V7S
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E, pH 6.2) | | 分子名称: | Matrix protein 1, PHOSPHATE ION | | 著者 | Musayev, F.N, Safo, M.K, Althufairi, B, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J, Zhou, Q. | | 登録日 | 2017-03-20 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
3GNE
 
 | | Crystal structure of alginate lyase vAL-1 from Chlorella virus | | 分子名称: | CITRATE ANION, GLYCEROL, VAL-1 | | 著者 | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | 登録日 | 2009-03-17 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
6I6B
 
 | |
6I6J
 
 | |
7A7B
 
 | | Bacillithiol Disulfide Reductase Bdr (YpdA) from Staphylococcus aureus | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YpdA family putative bacillithiol disulfide reductase Bdr | | 著者 | Hammerstad, M, Hersleth, H.-P. | | 登録日 | 2020-08-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase.
Biochemistry, 59, 2020
|
|
7A76
 
 | |
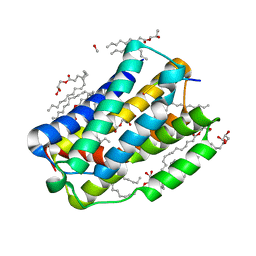
6I6H
 
 | |
6FTQ
 
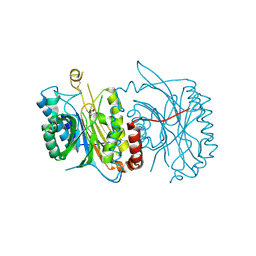 | |
7APR
 
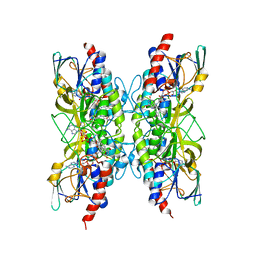 | | Bacillithiol Disulfide Reductase Bdr (YpdA) from Staphylococcus aureus | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YpdA family putative bacillithiol disulfide reductase Bdr | | 著者 | Hammerstad, M, Hersleth, H.-P. | | 登録日 | 2020-10-19 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase.
Biochemistry, 59, 2020
|
|
8JIH
 
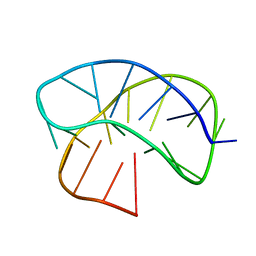 | |
8JIC
 
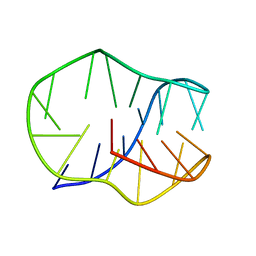 | |
2MX9
 
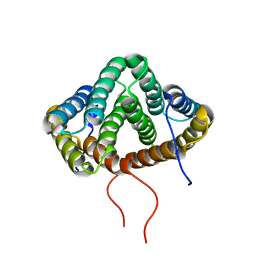 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 5.5 | | 分子名称: | Minor ampullate spidroin | | 著者 | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2MX8
 
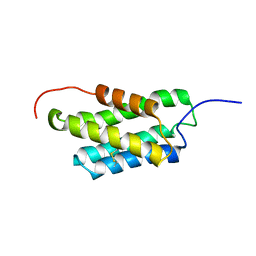 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 7.2 | | 分子名称: | Minor ampullate spidroin | | 著者 | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
6Q29
 
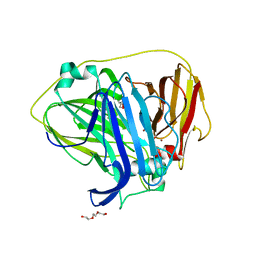 | |
6TYR
 
 | |
5GU7
 
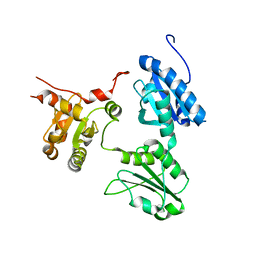 | | Crystal Structure of human ERp44 form II | | 分子名称: | Endoplasmic reticulum resident protein 44 | | 著者 | Watanabe, S, Inaba, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GU6
 
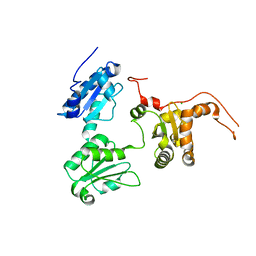 | | Crystal structure of Human ERp44 form I | | 分子名称: | CHLORIDE ION, Endoplasmic reticulum resident protein 44 | | 著者 | Watanabe, S, Inaba, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6W5S
 
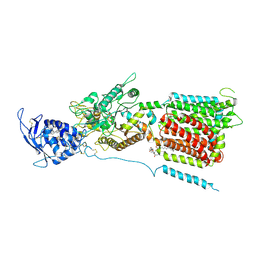 | | NPC1 structure in GDN micelles at pH 8.0 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5V
 
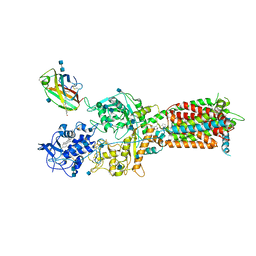 | | NPC1-NPC2 complex structure at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
7JWY
 
 | | Structure of SARS-CoV-2 spike at pH 4.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Kwong, P.D. | | 登録日 | 2020-08-26 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6W5R
 
 | | NPC1 structure in Nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5U
 
 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
