7EAZ
 
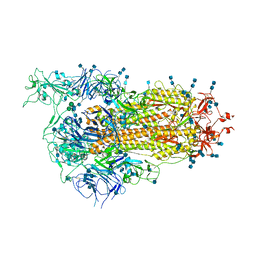 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2021-03-08 | | 公開日 | 2021-06-23 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
5T38
 
 | | Crystal Structure of the N-terminal domain of EvdMO1 with SAH bound | | 分子名称: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | McCulloch, K.M, Berndt, S, Yamakawa, I, Chen, Q, Loukachevitch, L.V, Starbird, C, Perry, N.A, Iverson, T.M. | | 登録日 | 2016-08-25 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1502 Å) | | 主引用文献 | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
4KN8
 
 | | Crystal structure of Bs-TpNPPase | | 分子名称: | Thermostable NPPase | | 著者 | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | 登録日 | 2013-05-09 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.502 Å) | | 主引用文献 | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
7EB5
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2021-03-08 | | 公開日 | 2021-06-23 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB3
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2021-03-08 | | 公開日 | 2021-06-23 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB4
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2021-03-08 | | 公開日 | 2021-06-23 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
2BED
 
 | | Structure of FPT bound to inhibitor SCH207736 | | 分子名称: | (11S)-8-CHLORO-11-[1-(METHYLSULFONYL)PIPERIDIN-4-YL]-6-PIPERAZIN-1-YL-11H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDINE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | 著者 | Strickland, C. | | 登録日 | 2005-10-24 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Enhanced FTase activity achieved via piperazine interaction with catalytic zinc.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8Z1G
 
 | | Cryo-EM structure of human ELAC2-pre-tRNA | | 分子名称: | Homo sapiens mitochondrion pre-tRNA-Tyr, PHOSPHATE ION, ZINC ION, ... | | 著者 | Liu, Z.M, Xue, C.Y. | | 登録日 | 2024-04-11 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into human ELAC2 as a tRNA 3' processing enzyme.
Nucleic Acids Res., 52, 2024
|
|
4JE3
 
 | |
8TOW
 
 | | Structure of a mutated photosystem II complex reveals perturbation of the oxygen-evolving complex | | 分子名称: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Flesher, D.A, Liu, J, Wang, J, Gisriel, C.J, Yang, K.R, Batista, V.S, Debus, R.J, Brudvig, G.W. | | 登録日 | 2023-08-04 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Mutation-induced shift of the photosystem II active site reveals insight into conserved water channels.
J.Biol.Chem., 300, 2024
|
|
7NER
 
 | |
5NNJ
 
 | |
7NES
 
 | |
2BOD
 
 | | Catalytic domain of endo-1,4-glucanase Cel6A from Thermobifida fusca in complex with methyl cellobiosyl-4-thio-beta-cellobioside | | 分子名称: | ENDOGLUCANASE E-2, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | 著者 | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | 登録日 | 2005-04-10 | | 公開日 | 2005-10-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
8QU7
 
 | |
7ENY
 
 | | Crystal structure of hydroxysteroid dehydrogenase from Escherichia coli | | 分子名称: | 7alpha-hydroxysteroid dehydrogenase | | 著者 | Kim, K.-H, Lee, C.W, Pardhe, D.P, Hwang, J, Do, H, Lee, Y.M, Lee, J.H, Oh, T.-J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Crystal structure of an apo 7 alpha-hydroxysteroid dehydrogenase reveals key structural changes induced by substrate and co-factor binding.
J.Steroid Biochem.Mol.Biol., 212, 2021
|
|
2J3X
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 3.0 (mut-S361R) | | 分子名称: | C2 TOXIN COMPONENT I, GLYCEROL, SULFATE ION | | 著者 | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | 登録日 | 2006-08-23 | | 公開日 | 2006-10-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2Z8L
 
 | | Crystal Structure of the Staphylococcal superantigen-like protein SSL5 at pH 4.6 complexed with sialyl Lewis X | | 分子名称: | Exotoxin 3, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Baker, H.M, Basu, I, Chung, M.C, Caradoc Davies, T, Fraser, J.D, Baker, E.N. | | 登録日 | 2007-09-06 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of the staphylococcal toxin SSL5 in complex with sialyl Lewis X reveal a conserved binding site that shares common features with viral and bacterial sialic acid binding proteins
J.Mol.Biol., 374, 2007
|
|
6VKD
 
 | | Crystal Structure of Inhibitor JNJ-36689282 in Complex with Prefusion RSV F Glycoprotein | | 分子名称: | 1-cyclopropyl-3-({1-[3-(methylsulfonyl)propyl]-1H-pyrrolo[3,2-c]pyridin-2-yl}methyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one, CHLORIDE ION, Prefusion RSV F (DS-Cav1), ... | | 著者 | McLellan, J.S. | | 登録日 | 2020-01-20 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
1ZB7
 
 | | Crystal Structure of Botulinum Neurotoxin Type G Light Chain | | 分子名称: | CITRATE ANION, ZINC ION, neurotoxin | | 著者 | Arndt, J.W, Yu, W, Bi, F, Stevens, R.C. | | 登録日 | 2005-04-07 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of botulinum neurotoxin type g light chain: serotype divergence in substrate recognition
Biochemistry, 44, 2005
|
|
3LFO
 
 | |
1ZJG
 
 | | 13mer-co | | 分子名称: | 5'-D(*AP*TP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*CP*A)-3', COBALT HEXAMMINE(III), ... | | 著者 | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | 登録日 | 2005-04-28 | | 公開日 | 2005-05-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
3BQ1
 
 | | Insertion ternary complex of Dbh DNA polymerase | | 分子名称: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*DGP*DAP*DAP*DGP*DCP*DCP*DGP*DGP*DCP*DG)-3'), ... | | 著者 | Pata, J.D, Wilson, R.C. | | 登録日 | 2007-12-19 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh.
Mol.Cell, 29, 2008
|
|
4JIU
 
 | | Crystal structure of the metallopeptidase zymogen of Pyrococcus abyssi abylysin | | 分子名称: | GLYCEROL, Proabylysin, ZINC ION | | 著者 | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | 登録日 | 2013-03-07 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
3VBC
 
 | |
