7LWF
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-9320 | | 分子名称: | B-cell lymphoma 6 protein, GLYCEROL, N-(3-chloropyridin-4-yl)-2-[5-(3-cyano-4-hydroxyphenyl)-3-methyl-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]acetamide | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2021-03-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LZS
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11029 | | 分子名称: | 3-chloro-5-{7-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
6OF7
 
 | | Crystal structure of the CRY1-PER2 complex | | 分子名称: | Cryptochrome-1, Period circadian protein homolog 2 | | 著者 | Michael, A.K, Fribourgh, J.L, Tripathi, S.M, Partch, C.L. | | 登録日 | 2019-03-28 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing.
Elife, 9, 2020
|
|
4YWC
 
 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | 分子名称: | Protein TIFY 7, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2015-03-20 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
5NYZ
 
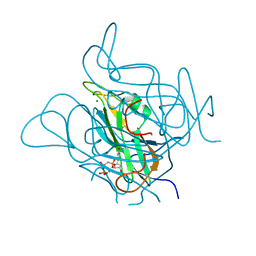 | | Twist and induce: Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 dUTPase. D95E mutant from dUTPase 80alpha phage. | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | 著者 | Alite, C, Humphrey, S, Donderis, J, Maiques, E, Ciges-Tomas, J.R, Penades, J.R, Marina, A. | | 登録日 | 2017-05-12 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 alpha dUTPase.
Sci Rep, 7, 2017
|
|
4RS9
 
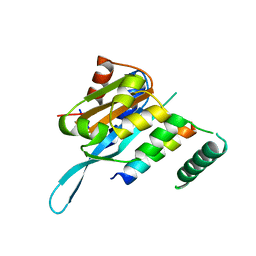 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | 分子名称: | Protein TIFY 7, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
8SVD
 
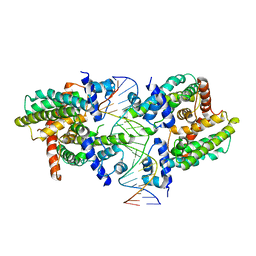 | |
8SSU
 
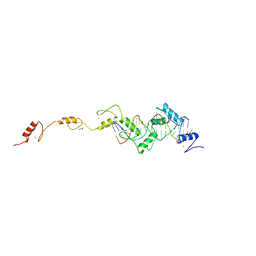 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SST
 
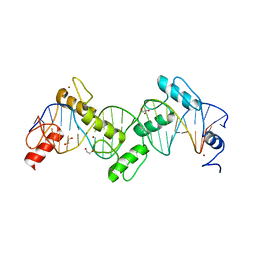 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) K365T Mutant Complexed with 23mer | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
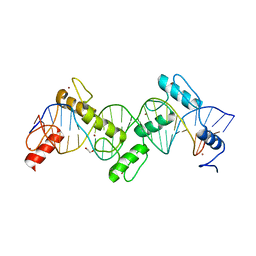 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSQ
 
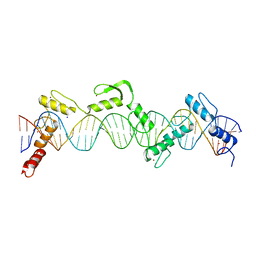 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | 分子名称: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
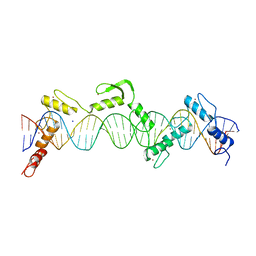 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | 分子名称: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
7WQU
 
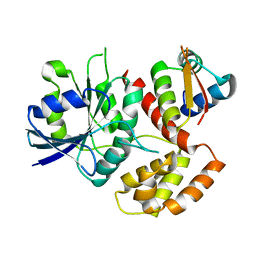 | | FeoC from Klebsiella pneumoniae | | 分子名称: | Ferrous iron transport protein B, Probable [Fe-S]-dependent transcriptional repressor | | 著者 | Hsueh, K.L, Yu, L.K, Hsieh, Y.C, Hsiao, Y.Y, Chen, C.J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (4.202 Å) | | 主引用文献 | FeoC from Klebsiella pneumoniae uses its iron sulfur cluster to regulate the GTPase activity of the ferrous iron channel.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
7PWJ
 
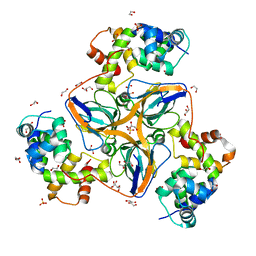 | | dUTPase from human in complex with Stl | | 分子名称: | 1,2-ETHANEDIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, ... | | 著者 | Sanz-Frasquet, C, Marina, A. | | 登録日 | 2021-10-06 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.944 Å) | | 主引用文献 | The Bacteriophage-Phage-Inducible Chromosomal Island Arms Race Designs an Interkingdom Inhibitor of dUTPases.
Microbiol Spectr, 11, 2023
|
|
2VXF
 
 | |
8CAM
 
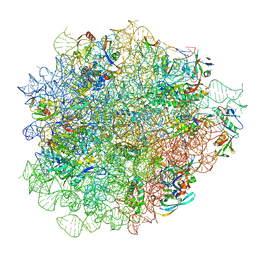 | | Evernimicin bound to the 50S subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | 著者 | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | 登録日 | 2023-01-24 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.86 Å) | | 主引用文献 | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGV
 
 | | Tiamulin bound to the 50S subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | 著者 | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | 登録日 | 2023-02-06 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.66 Å) | | 主引用文献 | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGK
 
 | | Lincomycin and Avilamycin bound to the 50S subunit | | 分子名称: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 23S rRNA, 50S ribosomal protein L15, ... | | 著者 | Paternoga, H, Crowe-McAuliffe, C, Beckert, B, Wilson, D.N. | | 登録日 | 2023-02-05 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.64 Å) | | 主引用文献 | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGD
 
 | | Clindamycin bound to the 50S subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | 著者 | Paternoga, H, Koller, T.O, Beckert, B, Wilson, D.N. | | 登録日 | 2023-02-03 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.98 Å) | | 主引用文献 | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CEU
 
 | | Retapamulin and Capreomycin bound to the 50S subunit | | 分子名称: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | 著者 | Paternoga, H, Beckert, B, Wilson, D.N. | | 登録日 | 2023-02-02 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.83 Å) | | 主引用文献 | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4WWW
 
 | | Crystal structure of the E. coli ribosome bound to CEM-101 | | 分子名称: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S rRNA, 23S rRNA, ... | | 著者 | Dunkle, J.A, Zhang, W, Cate, J.H.D, Mankin, A.S. | | 登録日 | 2014-11-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis.
Antimicrob. Agents Chemother., 54, 2010
|
|
4UV9
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | 分子名称: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | 著者 | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | 登録日 | 2014-08-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UVB
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1S,2R) | | 分子名称: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,7aS)-1-amino-1,10,11-trimethyl-4,6-dioxo-3-phenyl-2,3,5,6,7,7a-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | 著者 | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | 登録日 | 2014-08-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UXN
 
 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | 著者 | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | 登録日 | 2014-08-27 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
4UVC
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Phenyl-Tranylcypromine | | 分子名称: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-[5-[(1S)-1-azanyl-1,3-diphenyl-propyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-4aH-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | 著者 | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | 登録日 | 2014-08-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
