4FRC
 
 | |
7G1L
 
 | | Crystal Structure of human FABP4 in complex with 6-(1,3-benzodioxol-5-ylmethyl)-3-sulfanyl-1,2,4-triazin-5-ol | | 分子名称: | 6-[(2H-1,3-benzodioxol-5-yl)methyl]-3-sulfanyl-1,2,4-triazin-5-ol, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
6Z7I
 
 | |
6K9J
 
 | |
5RD8
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 30 | | 分子名称: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-06-17 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4FPT
 
 | | Carbonic Anhydrase II in complex with ethyl (2Z,4R)-2-(sulfamoylimino)-1,3-thiazolidine-4-carboxylate | | 分子名称: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Di Pizio, A, Heine, A, Klebe, G. | | 登録日 | 2012-06-22 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | High resolution crystal structures of Carbonic Anhydrase II in complex with novel sulfamide binders
To be Published
|
|
4FU5
 
 | |
3I5E
 
 | |
5HMV
 
 | | Re refinement of 4mwk. | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | 著者 | Helliwell, J.R. | | 登録日 | 2016-01-17 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn, 3, 2016
|
|
5RDX
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 54 | | 分子名称: | Endothiapepsin | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-06-17 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | 分子名称: | Computational design, MID1-apo1 | | 著者 | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
5RVH
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000265642 | | 分子名称: | Non-structural protein 3, quinoline-3-carboxylic acid | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6LIP
 
 | |
4KXU
 
 | | Human transketolase in covalent complex with donor ketose D-fructose-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, D-SORBITOL-6-PHOSPHATE, MAGNESIUM ION, ... | | 著者 | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | 登録日 | 2013-05-28 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
5KXV
 
 | | Structure Proteinase K at 0.98 Angstroms | | 分子名称: | CALCIUM ION, GLYCEROL, NITRATE ION, ... | | 著者 | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | 登録日 | 2016-07-20 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
5XR0
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20T mutant (NFE6, AFP), P21 form | | 分子名称: | Ice-structuring protein | | 著者 | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | 登録日 | 2017-06-07 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7YZ7
 
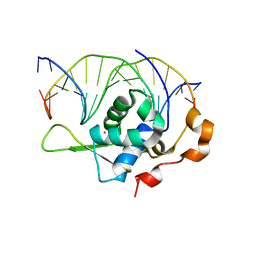 | | Crystal structure of the zebrafish FoxH1 bound to the TGTGGATT site | | 分子名称: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | 著者 | Pluta, R, Macias, M.J. | | 登録日 | 2022-02-18 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
6TX6
 
 | |
2G58
 
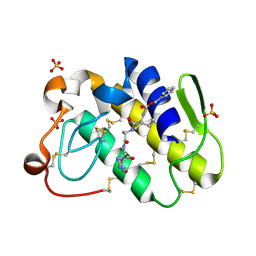 | | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution | | 分子名称: | (PHQ)IARS, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | 著者 | Prem Kumar, R, Singh, N, Somvanshi, R.K, Ethayathulla, A.S, Dey, S, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | 登録日 | 2006-02-22 | | 公開日 | 2006-03-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution
To be Published
|
|
2ZPM
 
 | |
1IXH
 
 | |
2BZZ
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | 分子名称: | ACETIC ACID, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, NONSECRETORY RIBONUCLEASE | | 著者 | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | 登録日 | 2005-08-24 | | 公開日 | 2006-01-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
4QBX
 
 | |
4WPK
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form I | | 分子名称: | CITRIC ACID, SODIUM ION, Uracil-DNA glycosylase | | 著者 | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | 登録日 | 2014-10-20 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7OYN
 
 | | Carbonic anhydrase II in complex with Hit3 (MH57) | | 分子名称: | Carbonic anhydrase 2, Hit3 (MH57), ZINC ION | | 著者 | Kugler, M, Brynda, J, Rezacova, P. | | 登録日 | 2021-06-24 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
