1FL8
 
 | |
7B9V
 
 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | 分子名称: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | 著者 | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | 登録日 | 2020-12-14 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
6JM9
 
 | |
7WBX
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBV
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | 分子名称: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
3PHC
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase in complex with DADMe-ImmG | | 分子名称: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Ho, M, Edwards, A.A, Almo, S.C, Schramm, V.L. | | 登録日 | 2010-11-03 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase in complex with DADMe-ImmG
To be Published
|
|
6NQB
 
 | |
4GMH
 
 | | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase | | 分子名称: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ACETATE ION | | 著者 | Brown, R.L, Anderson, B.F, Norris, G.E, Tyler, P.C, Evans, G.B, Sutherland-Smith, A.J. | | 登録日 | 2012-08-15 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase
To be Published
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
2YKN
 
 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with a Difluoromethylbenzoxazole (DFMB) Pyrimidine Thioether derivative, a non-nucleoside RT inhibitor (NNRTI) | | 分子名称: | 2-[DIFLUORO-[(4-METHYL-PYRIMIDINYL)-THIO]METHYL]-BENZOXAZOLE, CALCIUM ION, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | 著者 | Boyer, J, Arnoult, E, Medebielle, M, Guillemont, J, Unge, T, Unge, J, Jochmans, D. | | 登録日 | 2011-05-28 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Difluoromethylbenzoxazole Pyrimidine Thioether Derivatives: A Novel Class of Potent Non-Nucleoside HIV-1 Reverse Transcriptase Inhibitors.
J.Med.Chem., 54, 2011
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
2YKM
 
 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with a Difluoromethylbenzoxazole (DFMB) Pyrimidine Thioether derivative, a non-nucleoside RT inhibitor (NNRTI) | | 分子名称: | 2-[DIFLUORO-[(4-METHYL-PYRIMIDINYL)-THIO]METHYL]-BENZOXAZOLE, CALCIUM ION, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | 著者 | Boyer, J, Arnoult, E, Medebielle, M, Guillemont, J, Unge, T, Unge, J, Jochmans, D. | | 登録日 | 2011-05-28 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Difluoromethylbenzoxazole Pyrimidine Thioether Derivatives: A Novel Class of Potent Non-Nucleoside HIV-1 Reverse Transcriptase Inhibitors.
J.Med.Chem., 54, 2011
|
|
8JH2
 
 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | 著者 | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2023-05-22 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH3
 
 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2023-05-22 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
3PNP
 
 | |
2H8G
 
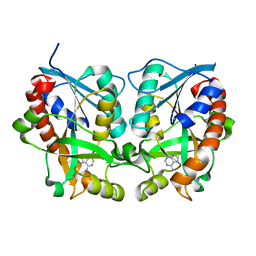 | | 5'-Methylthioadenosine Nucleosidase from Arabidopsis thaliana | | 分子名称: | 5'-Methylthioadenosine Nucleosidase, ADENINE | | 著者 | Park, E.Y, Oh, S.I, Nam, M.J, Shin, J.S, Kim, K.N, Song, H.K. | | 登録日 | 2006-06-07 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-A resolution
Proteins, 65, 2006
|
|
3AN2
 
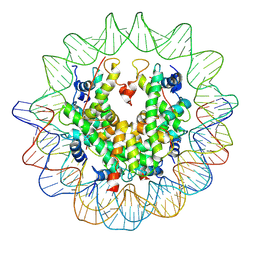 | | The structure of the centromeric nucleosome containing CENP-A | | 分子名称: | 147 mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Tachiwana, H, Kagawa, W, Shiga, T, Saito, K, Osakabe, A, Hayashi-Takanaka, Y, Park, S.-Y, Kimura, H, Kurumizaka, H. | | 登録日 | 2010-08-27 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structure of the human centromeric nucleosome containing CENP-A
Nature, 476, 2011
|
|
4BMX
 
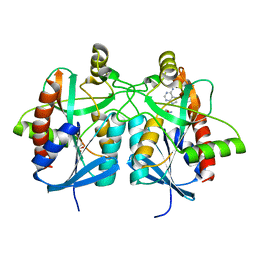 | | Native structure of futalosine hydrolase of Helicobacter pylori strain 26695 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, MTA/SAH NUCLEOSIDASE | | 著者 | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | 登録日 | 2013-05-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9B1E
 
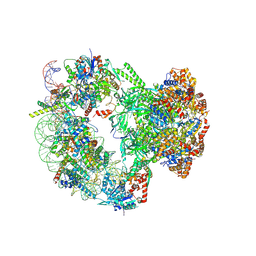 | | Cryo-EM structure of native SWR1 bound to nucleosome (composite structure) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Louder, R.K, Park, G, Wu, C. | | 登録日 | 2024-03-13 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Molecular basis of global promoter sensing and nucleosome capture by the SWR1 chromatin remodeler.
Cell, 187, 2024
|
|
7VDV
 
 | | The overall structure of human chromatin remodeling PBAF-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2,AT-rich interactive domain-containing protein 2, Actin, ... | | 著者 | Chen, Z.C, Chen, K.J, Yuan, J.J. | | 登録日 | 2021-09-07 | | 公開日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
7VDT
 
 | | The motor-nucleosome module of human chromatin remodeling PBAF-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (207-MER), ... | | 著者 | Chen, Z.C, Chen, K.J, Yuan, J.J. | | 登録日 | 2021-09-07 | | 公開日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
7B9C
 
 | |
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
