1VSH
 
 | |
1VSJ
 
 | |
1VSI
 
 | |
8SF7
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain MEC17 | | 分子名称: | CFAM166A, CFAM166B, CFAM166C, ... | | 著者 | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | 登録日 | 2023-04-10 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Effect of alpha-tubulin acetylation on the doublet microtubule structure.
Elife, 12, 2024
|
|
8FS4
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
3ST1
 
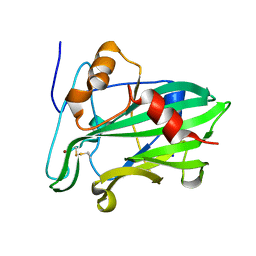 | | Crystal structure of Necrosis and Ethylene inducing Protein 2 from the causal agent of cocoa's Witches Broom disease | | 分子名称: | Necrosis-and ethylene-inducing protein, SODIUM ION, ZINC ION | | 著者 | Oliveira, J.F, Zaparoli, G, Barsottini, M.R.O, Ambrosio, A.L.B, Pereira, G.A.G, Dias, S.M.G. | | 登録日 | 2011-07-08 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Crystal Structure of Necrosis- and Ethylene-Inducing Protein 2 from the Causal Agent of Cacao's Witches' Broom Disease Reveals Key Elements for Its Activity.
Biochemistry, 50, 2011
|
|
1DXP
 
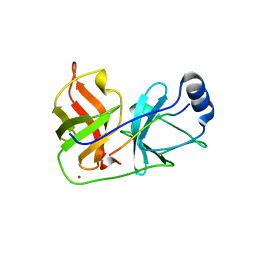 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | 分子名称: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | 著者 | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | 登録日 | 2000-01-13 | | 公開日 | 2001-01-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
8FS3
 
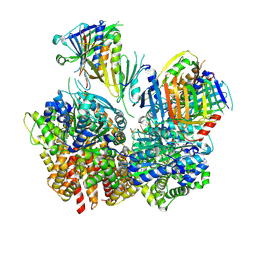 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
1CYN
 
 | |
1VOK
 
 | |
4BFP
 
 | | Crystal structure of human tankyrase 2 in complex with WIKI4 | | 分子名称: | 2-[3-[[4-(4-methoxyphenyl)-5-pyridin-4-yl-1,2,4-triazol-3-yl]sulfanyl]propyl]benzo[de]isoquinoline-1,3-dione, SULFATE ION, TANKYRASE-2, ... | | 著者 | Haikarainen, T, Narwal, M, Lehtio, L. | | 登録日 | 2013-03-21 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis and Selectivity of Tankyrase Inhibition by a Wnt Signaling Inhibitor Wiki4
Plos One, 8, 2013
|
|
3MHK
 
 | | Human tankyrase 2 - catalytic PARP domain in complex with 2-(2-pyridyl)-7,8-dihydro-5h-thiino[4,3-d]pyrimidin-4-ol | | 分子名称: | 2-pyridin-2-yl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, Tankyrase-2, ... | | 著者 | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-04-08 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
3MHJ
 
 | | Human tankyrase 2 - catalytic PARP domain in complex with 1-methyl-3-(trifluoromethyl)-5h-benzo[c][1,8]naphtyridine-6-one | | 分子名称: | 1-methyl-3-(trifluoromethyl)benzo[c][1,8]naphthyridin-6(5H)-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-04-08 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
4J1Z
 
 | |
1CKI
 
 | |
2JX3
 
 | |
7KRD
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-(3-chloro-5-cyanophenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ702) | | 分子名称: | 4-(3-chloro-5-cyanophenoxy)-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | 著者 | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-11-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.91 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
7U5Z
 
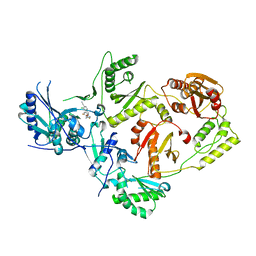 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with JLJ353 | | 分子名称: | 2-chloro-4-({5-[(2,6-difluorophenyl)methyl]-1,3-oxazol-2-yl}amino)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2022-03-03 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design, synthesis, and biological testing of biphenylmethyloxazole inhibitors targeting HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
6WPH
 
 | |
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
