8BX3
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074372) | | 分子名称: | 14-3-3 protein sigma, 2-(4-bromanylphenoxy)-~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-2-methyl-propanamide, ERalpha peptide, ... | | 著者 | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | 登録日 | 2022-12-07 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4YO5
 
 | | EAEC T6SS TssA-Cterminus | | 分子名称: | TssA | | 著者 | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | 登録日 | 2015-03-11 | | 公開日 | 2016-02-17 | | 最終更新日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
8BXI
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074361) | | 分子名称: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | 著者 | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | 登録日 | 2022-12-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BXN
 
 | |
8BYD
 
 | |
8BWX
 
 | |
8BX0
 
 | |
5D3G
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | 分子名称: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | 登録日 | 2015-08-06 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
8BYC
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1075316) | | 分子名称: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | 著者 | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | 登録日 | 2022-12-12 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BXM
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074397) | | 分子名称: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | 著者 | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | 登録日 | 2022-12-09 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5JGW
 
 | | Crystal structure of maize AKR4C13 in complex with NADP and acetate | | 分子名称: | ACETATE ION, Aldose reductase, AKR4C13, ... | | 著者 | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | 登録日 | 2016-04-20 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of maize AKR4C13 in complex with NADP and acetate
To Be Published
|
|
1GEN
 
 | | C-TERMINAL DOMAIN OF GELATINASE A | | 分子名称: | CALCIUM ION, CHLORIDE ION, GELATINASE A, ... | | 著者 | Libson, A.M, Gittis, A.G, Collier, I.E, Marmer, B.L, Goldberg, G.G, Lattman, E.E. | | 登録日 | 1995-07-19 | | 公開日 | 1996-08-17 | | 最終更新日 | 2018-03-21 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Nat.Struct.Biol., 2, 1995
|
|
5JH7
 
 | | Tubulin-Eribulin complex | | 分子名称: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Doodhi, H, Prota, A.E, Rodriguez-Garcia, R, Xiao, H, Custar, D.W, Bargsten, K, Katrukha, E.A, Hilbert, M, Hua, S, Jiang, K, Grigoriev, I, Yang, C.-P.H, Cox, D, Band Horwitz, S, Kapitein, L.C, Akhmanova, A, Steinmetz, M.O. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes.
Curr.Biol., 26, 2016
|
|
7PIK
 
 | | Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon | | 分子名称: | Right end fragment of Tn7 transposon, Transposon Tn7 transposition protein TnsB | | 著者 | Kaczmarska, Z, Czarnocki-Cieciura, M, Rawski, M, Nowotny, M. | | 登録日 | 2021-08-20 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Mol.Cell, 82, 2022
|
|
8BYG
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1047648) | | 分子名称: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[2-[(2-carbamimidoyl-1-benzothiophen-4-yl)-methyl-amino]ethyl]-2-(4-chloranylphenoxy)-~{N},2-dimethyl-propanamide | | 著者 | Visser, E.J, Sijbesma, E, Ottmann, C. | | 登録日 | 2022-12-12 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7OB1
 
 | | OLIGOPEPTIDASE B FROM S. PROTEOMACULANS WITH MODIFIED HINGE | | 分子名称: | Oligopeptidase B, SPERMINE | | 著者 | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Korzhenevskiy, D.A, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | 登録日 | 2021-04-20 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
5JGY
 
 | | Crystal structure of maize AKR4C13 in P21 space group | | 分子名称: | 1,2-ETHANEDIOL, 4-{[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]oxy}butanoic acid (non-preferred name), Aldose reductase, ... | | 著者 | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | 登録日 | 2016-04-20 | | 公開日 | 2017-05-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of maize AKR4C13 in P21 space group
To Be Published
|
|
8EQJ
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQT
 
 | | Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-09 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
6QG9
 
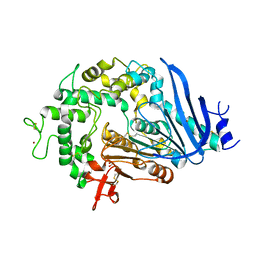 | | Crystal structure of Ideonella sakaiensis MHETase | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
8EQS
 
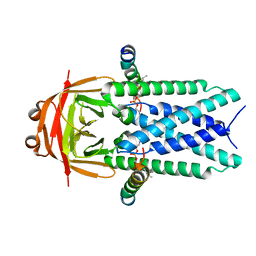 | | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-09 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
7OQR
 
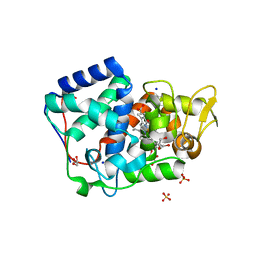 | | Crystal structure of Trypanosoma cruzi peroxidase | | 分子名称: | ACETATE ION, Ascorbate peroxidase, GLYCEROL, ... | | 著者 | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | 登録日 | 2021-06-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | 分子名称: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-04-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
7UPL
 
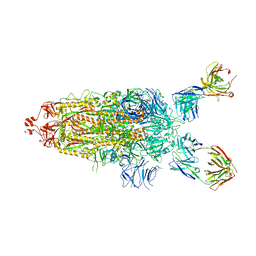 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Patel, A, Ortlund, E. | | 登録日 | 2022-04-15 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
6MK7
 
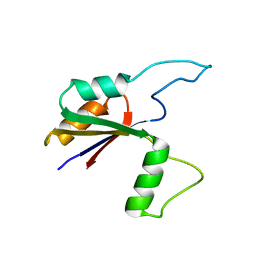 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | 分子名称: | Cell division protein FtsX | | 著者 | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | 登録日 | 2018-09-25 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
