7BN8
 
 | | Notum fragment_3 (4H,5H-naphtho[1,2-b]thiophene-2-carboxylic acid) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4H,5H-naphtho[1,2-b]thiophene-2-carboxylic acid, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2021-01-21 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Notum Inhibitor
To Be Published
|
|
9EV5
 
 | | Corynebacterium glutamicum CS176 pyruvate:quinone oxidoreductase (PQO) in complex with FAD and thiamine diphosphate-magnesium ion | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | 著者 | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | 登録日 | 2024-03-28 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.863 Å) | | 主引用文献 | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 2024
|
|
6D51
 
 | | Crystal structure of L,D-transpeptidase 3 from Mycobacterium tuberculosis in complex with a faropenem-derived adduct | | 分子名称: | ACETYL GROUP, CALCIUM ION, Probable L,D-transpeptidase 3 | | 著者 | Libreros, G.A, Dias, M.V.B. | | 登録日 | 2018-04-19 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural Basis for the Interaction and Processing of beta-Lactam Antibiotics by l,d-Transpeptidase 3 (LdtMt3) from Mycobacterium tuberculosis.
ACS Infect Dis, 5, 2019
|
|
4UCZ
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | 分子名称: | GUANOSINE, GUANOSINE-5'-TRIPHOSPHATE, RNA-DIRECTED RNA POLYMERASE L, ... | | 著者 | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | 登録日 | 2014-12-05 | | 公開日 | 2015-11-18 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
7M67
 
 | |
7M73
 
 | |
7M79
 
 | |
6DGA
 
 | |
6DIZ
 
 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | 分子名称: | SPHINGOSINE, VP1, VP2, ... | | 著者 | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | 登録日 | 2018-05-24 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
4UB7
 
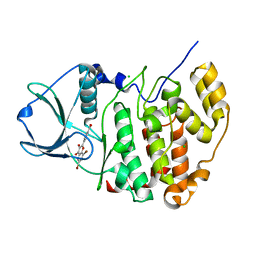 | | High-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) showing an extreme distortion of the ATP-binding loop combined with a pi-halogen bond | | 分子名称: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | 著者 | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | 登録日 | 2014-08-12 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
9ASS
 
 | |
6DCD
 
 | |
4U5A
 
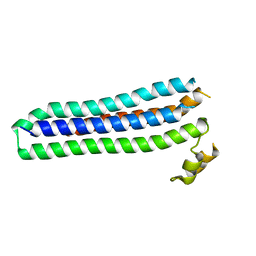 | | Sporozoite Protein for Cell Traversal | | 分子名称: | Sporozoite microneme protein essential for cell traversal | | 著者 | Hamaoka, B.Y. | | 登録日 | 2014-07-25 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of the Essential Plasmodium Host Cell Traversal Protein SPECT1.
Plos One, 9, 2014
|
|
6DML
 
 | | A multiconformer ligand model of 3,5 dimethylisoxaxole bound to the bromodomain of human BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 4-((2-(tert-butyl)phenyl)amino)-7-(3,5-dimethylisoxazol-4-yl)-6-methoxy-1,5-naphthyridine-3-carboxylic acid, Bromodomain-containing protein 4 | | 著者 | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | 登録日 | 2018-06-05 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DMS
 
 | | Endo-fucoidan hydrolase MfFcnA4_H294Q from glycoside hydrolase family 107 | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | 著者 | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | 登録日 | 2018-06-05 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
4UG0
 
 | | STRUCTURE OF THE HUMAN 80S RIBOSOME | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S RIBOSOMAL PROTEIN, ... | | 著者 | Khatter, H, Myasnikov, A.G, Natchiar, S.K, Klaholz, B.P. | | 登録日 | 2015-03-20 | | 公開日 | 2015-06-10 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of the human 80S ribosome
NATURE, 520, 2015
|
|
6DLG
 
 | | Crystal structure of a SHIP1 surface entropy reduction mutant | | 分子名称: | ISOPROPYL ALCOHOL, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Gardill, B.R, Cheung, S.T, Mui, A.L, Van Petegem, F. | | 登録日 | 2018-06-01 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Interleukin-10 and Small Molecule SHIP1 Allosteric Regulators Trigger Anti-Inflammatory Effects Through SHIP1/STAT3 Complexes
Biorxiv, 2020
|
|
4U91
 
 | | GephE in complex with Para-Phenyl crosslinked Glycine receptor beta subunit derived dimeric peptide | | 分子名称: | 1,1'-benzene-1,4-diylbis(1H-pyrrole-2,5-dione), 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | 登録日 | 2014-08-05 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Synthesis of High-Affinity Dimeric Inhibitors Targeting the Interactions between Gephyrin and Inhibitory Neurotransmitter Receptors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4UCY
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | 分子名称: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | 著者 | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | 登録日 | 2014-12-05 | | 公開日 | 2015-11-18 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
7M7C
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | 分子名称: | Carboxylesterase A | | 著者 | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | 登録日 | 2021-03-27 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
4UE5
 
 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | 分子名称: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | 著者 | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | 登録日 | 2014-12-15 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNI
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | 分子名称: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | 登録日 | 2014-05-28 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
6DMJ
 
 | | A multiconformer ligand model of inhibitor 53W bound to CREB binding protein bromodomain | | 分子名称: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[2-(4-methoxyphenyl)ethyl]-1-[2-(morpholin-4-yl)ethyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | 著者 | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | 登録日 | 2018-06-05 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
7MN2
 
 | | Rules for designing protein fold switches and their implications for the folding code | | 分子名称: | Sb2 | | 著者 | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | 登録日 | 2021-04-30 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
