9EM1
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | 分子名称: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | 著者 | Brenner, M, Gohla, A, Schindelin, H. | | 登録日 | 2024-03-07 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
9B59
 
 | |
9B5A
 
 | |
9B5H
 
 | |
9B5L
 
 | |
9B5X
 
 | |
9B5V
 
 | |
9CBM
 
 | | Cryo-EM structure of dexmedetomidine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein | | 分子名称: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Endolysin,Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Lou, J.S, Su, M, Wang, J, Do, H.N, Miao, Y, Huang, X.Y. | | 登録日 | 2024-06-19 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Distinct binding conformations of epinephrine with alpha- and beta-adrenergic receptors.
Exp.Mol.Med., 2024
|
|
9IKC
 
 | | Orf virus scaffolding protein Orfv075 | | 分子名称: | 62 kDa protein | | 著者 | Hyun, J, Kim, S, Ko, S, Kim, M, Jang, Y. | | 登録日 | 2024-06-27 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (1.9 Å) | | 主引用文献 | Cryo-EM structure of orf virus scaffolding protein orfv075.
Biochem.Biophys.Res.Commun., 728, 2024
|
|
9EX1
 
 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure B) | | 分子名称: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex | | 著者 | Tito, G, Merlino, A, Ferraro, G. | | 登録日 | 2024-04-05 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.785 Å) | | 主引用文献 | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9CBL
 
 | | Cryo-EM structure of epinephrine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein | | 分子名称: | Endolysin,Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Lou, J.S, Su, M, Wang, J, Do, H.N, Miao, Y, Huang, X.Y. | | 登録日 | 2024-06-19 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Distinct binding conformations of epinephrine with alpha- and beta-adrenergic receptors.
Exp.Mol.Med., 2024
|
|
9ATC
 
 | | ATCASE Y165F MUTANT | | 分子名称: | ASPARTATE TRANSCARBAMOYLASE, ZINC ION | | 著者 | Ha, Y, Allewell, N.M. | | 登録日 | 1998-06-26 | | 公開日 | 1999-07-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Intersubunit hydrogen bond acts as a global molecular switch in Escherichia coli aspartate transcarbamoylase.
Proteins, 33, 1998
|
|
9EX2
 
 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure C) | | 分子名称: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | 著者 | Tito, G, Merlino, A, Ferraro, G. | | 登録日 | 2024-04-05 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.172 Å) | | 主引用文献 | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9EX0
 
 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure A) | | 分子名称: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | 著者 | Tito, G, Merlino, A, Ferraro, G. | | 登録日 | 2024-04-05 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7T1M
 
 | |
7T1S
 
 | |
9AX6
 
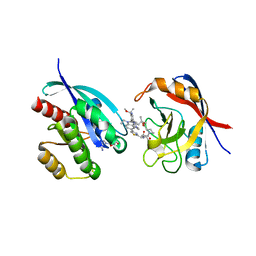 | | Tricomplex of RMC-6236, KRAS G12D, and CypA | | 分子名称: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | 著者 | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | 登録日 | 2024-03-05 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9G7H
 
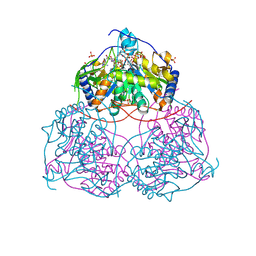 | | Human Sirt6 in complex with ADP-ribose and the inhibitor 2-Pr | | 分子名称: | 2,4-bis(oxidanylidene)-1-[2-oxidanylidene-2-[[(2S)-3-oxidanylidene-3-(propylamino)-2-[3-(3-pyridin-3-yl-1,2,4-oxadiazol-5-yl)propanoylamino]propyl]amino]ethyl]pyrimidine-5-carboxamide, NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, ... | | 著者 | You, W, Steegborn, C. | | 登録日 | 2024-07-21 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Elucidating the Unconventional Binding Mode of a DNA-Encoded Library Hit Provides a Blueprint for Sirtuin 6 Inhibitor Development.
Chemmedchem, 2024
|
|
9B34
 
 | |
9B5G
 
 | |
7TB9
 
 | | Structural characterization of the biological synthetic peptide pCEMP1 | | 分子名称: | CEMP1-p1 | | 著者 | Lopez Giraldo, A, del Rio Portilla, F, Nidome Campos, M, Romo Arevalo, E, Arzate, H. | | 登録日 | 2021-12-21 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of cementum protein 1 derived peptide (CEMP1-p1) and its role in the mineralization process.
J.Pept.Sci., 29, 2023
|
|
3SY7
 
 | |
7NVO
 
 | | Human TRiC complex in open state with nanobody bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | 登録日 | 2021-03-15 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVN
 
 | | Human TRiC complex in closed state with nanobody and tubulin bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | 登録日 | 2021-03-15 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | 分子名称: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | 著者 | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | 登録日 | 2021-03-17 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
