6ZZY
 
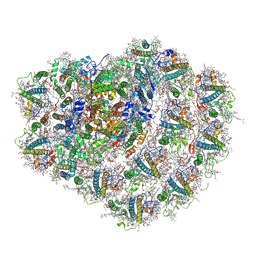 | | Structure of high-light grown Chlorella ohadii photosystem I | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6ZZX
 
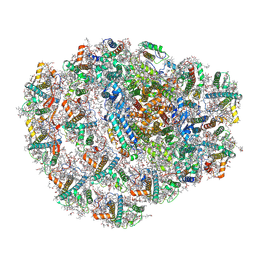 | | Structure of low-light grown Chlorella ohadii Photosystem I | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
5QGI
 
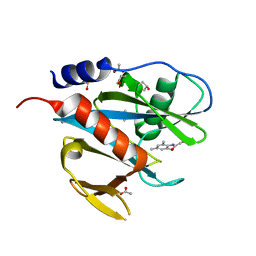 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000710a | | 分子名称: | 2-methoxy-~{N}-(2,4,6-trimethylphenyl)ethanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGT
 
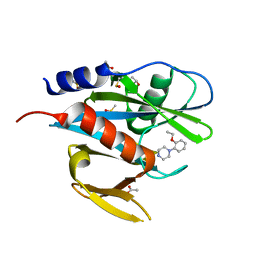 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000609a | | 分子名称: | 1-(2-ethoxyphenyl)piperazine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QH8
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with PCM-0102558 | | 分子名称: | 1-[(2S)-2-(4-methoxyphenyl)-2,3,6,7-tetrahydro-1H-azepin-1-yl]ethan-1-one, ACETATE ION, Peroxisomal coenzyme A diphosphatase NUDT7 | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
6G4G
 
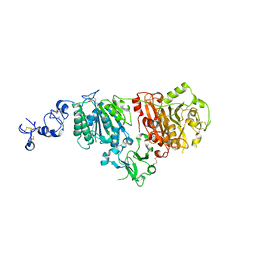 | | Full length ectodomain of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) including the SMB domains but with a partially disordered active site structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Dohler, C, Zebisch, M, Strater, N. | | 登録日 | 2018-03-27 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallization of ectonucleotide phosphodiesterase/pyrophosphatase-3 and orientation of the SMB domains in the full-length ectodomain.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5QGJ
 
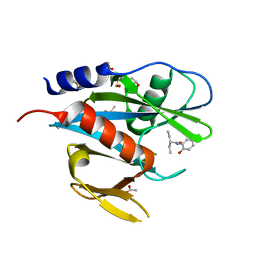 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000706a | | 分子名称: | 2-ethyl-N-(2-hydroxyphenyl)butanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGW
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000154 | | 分子名称: | 2-(4-methoxyphenyl)-N-(pyridin-3-yl)acetamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHC
 
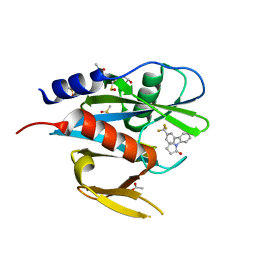 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with RK4-337 | | 分子名称: | 1-[3'-(trifluoromethyl)[1,1'-biphenyl]-2-yl]-1,3-dihydro-2H-pyrrol-2-one, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGV
 
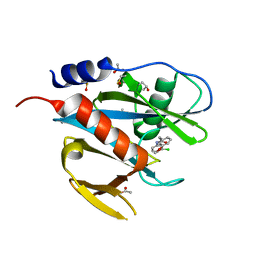 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with UNUYB062989 | | 分子名称: | ACETATE ION, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-2-(3-methoxyphenyl)acetamide, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QH9
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with PCM-0102716 | | 分子名称: | 1-[(2S)-2-(3-methoxyphenyl)pyrrolidin-1-yl]ethan-1-one, ACETATE ION, Peroxisomal coenzyme A diphosphatase NUDT7 | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4Y34
 
 | | Crystal Structure of Coxsackievirus B3 3D polymerase in complex with GPC-N143 | | 分子名称: | 2,2'-[(4-fluorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | 著者 | Vives-Adrian, L, Ferrer-Orta, C, Cerdaguer, N. | | 登録日 | 2015-02-10 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
4Y3C
 
 | |
4Y2C
 
 | |
6G16
 
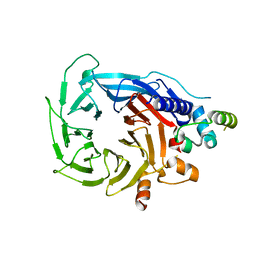 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | 分子名称: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | 著者 | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | 登録日 | 2018-03-20 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
6HHQ
 
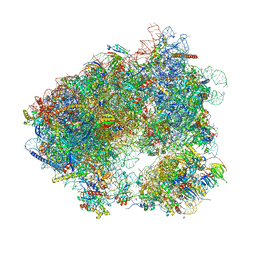 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | 分子名称: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | 登録日 | 2018-08-28 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.10000038 Å) | | 主引用文献 | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
1RK7
 
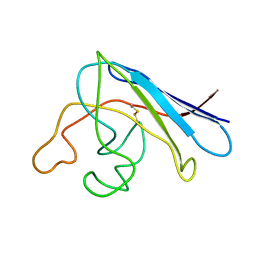 | | Solution structure of apo Cu,Zn Superoxide Dismutase: role of metal ions in protein folding | | 分子名称: | Superoxide dismutase [Cu-Zn] | | 著者 | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | 登録日 | 2003-11-21 | | 公開日 | 2003-12-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding
Biochemistry, 42, 2003
|
|
7TN6
 
 | |
4X8M
 
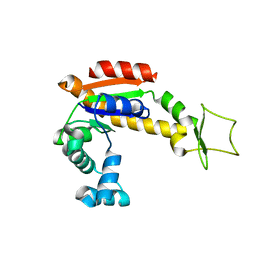 | | Crystal structure of E. coli Adenylate kinase Y171W mutant | | 分子名称: | Adenylate kinase | | 著者 | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | 登録日 | 2014-12-10 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
7TN5
 
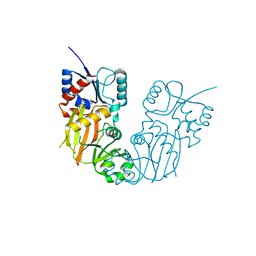 | |
7TN8
 
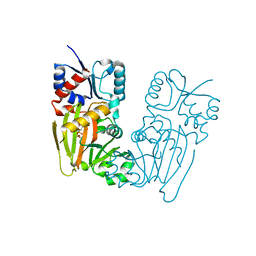 | |
7TN3
 
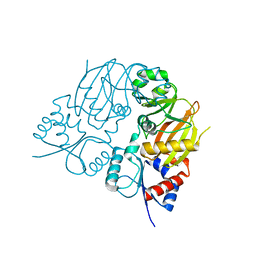 | |
7TN7
 
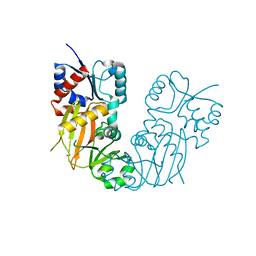 | |
3B0C
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form I | | 分子名称: | CITRIC ACID, Centromere protein T, Centromere protein W | | 著者 | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | 登録日 | 2011-06-08 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
6YSS
 
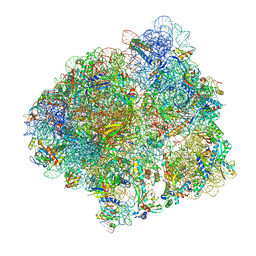 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | 登録日 | 2020-04-23 | | 公開日 | 2020-08-19 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
