4FLQ
 
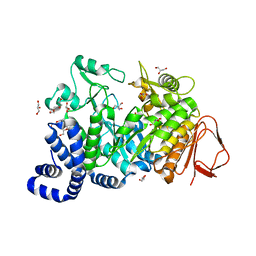 | | Crystal structure of Amylosucrase double mutant A289P-F290I from Neisseria polysaccharea. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | 著者 | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | 登録日 | 2012-06-15 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FEA
 
 | |
2FU3
 
 | |
1MKY
 
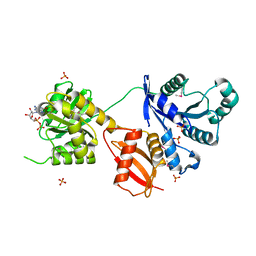 | | Structural Analysis of the Domain Interactions in Der, a Switch Protein Containing Two GTPase Domains | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Probable GTP-binding protein engA | | 著者 | Robinson, V.L, Hwang, J, Fox, E, Inouye, M, Stock, A.M. | | 登録日 | 2002-08-29 | | 公開日 | 2003-01-14 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Domain Arrangement of Der, a Switch Protein Containing Two GTPase Domains
Structure, 10, 2002
|
|
4FHD
 
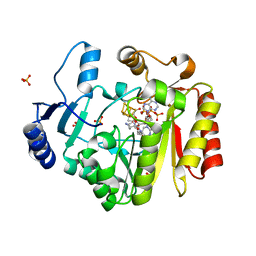 | | Spore photoproduct lyase complexed with dinucleoside spore photoproduct | | 分子名称: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, PYROPHOSPHATE 2-, ... | | 著者 | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | 登録日 | 2012-06-06 | | 公開日 | 2012-07-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
2FSG
 
 | |
1MOW
 
 | | E-DreI | | 分子名称: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*G)-3', 5'-D(*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', GLYCEROL, ... | | 著者 | Chevalier, B.S, Kortemme, T, Chadsey, M.S, Baker, D, Monnat Jr, R.J, Stoddard, B.L. | | 登録日 | 2002-09-10 | | 公開日 | 2002-11-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design, Activity and Structure of a Highly Specific Artificial Endonuclease
Mol.Cell, 10, 2002
|
|
4F9B
 
 | | Human CDC7 kinase in complex with DBF4 and PHA767491 | | 分子名称: | 2-(pyridin-4-yl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Cell division cycle 7-related protein kinase, Protein DBF4 homolog A, ... | | 著者 | Hughes, S, Cherepanov, P. | | 登録日 | 2012-05-18 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of human CDC7 kinase in complex with its activator DBF4.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FAB
 
 | | THREE-DIMENSIONAL STRUCTURE OF A FLUORESCEIN-FAB COMPLEX CRYSTALLIZED IN 2-METHYL-2,4-PENTANEDIOL | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, IGG2A-KAPPA 4-4-20 FAB (HEAVY CHAIN), ... | | 著者 | Herron, J.N, He, X, Mason, M.L, Vossjunior, E.W, Edmundson, A.B. | | 登録日 | 1989-04-10 | | 公開日 | 1990-07-15 | | 最終更新日 | 2017-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Three-dimensional structure of a fluorescein-Fab complex crystallized in 2-methyl-2,4-pentanediol.
Proteins, 5, 1989
|
|
4FAK
 
 | | Crystal Structure of OrfX in Complex with S-Adenosylmethionine | | 分子名称: | PHOSPHATE ION, Ribosomal RNA large subunit methyltransferase H, S-ADENOSYLMETHIONINE, ... | | 著者 | Safo, M.K, Musayev, F.N, Boundy, S, Archer, G.L, Rife, J.P, O'Farrell, H.C. | | 登録日 | 2012-05-22 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Characterization of the Staphylococcus aureus rRNA Methyltransferase Encoded by orfX, the Gene Containing the Staphylococcal Chromosome Cassette mec (SCCmec) Insertion Site.
J.Biol.Chem., 288, 2013
|
|
2G09
 
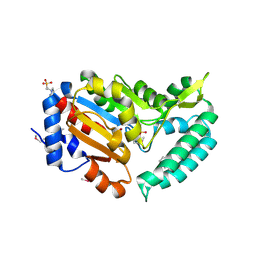 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product complex | | 分子名称: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-02-11 | | 公開日 | 2006-04-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
7RK8
 
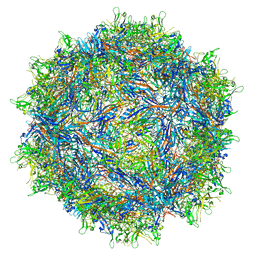 | |
7RA4
 
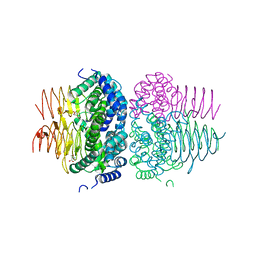 | | Crystal structure of Neisseria gonorrhoeae serine acetyltransferase (CysE) in complex with serine | | 分子名称: | SERINE, Serine acetyltransferase | | 著者 | Hicks, J.L, Oldham, K.E, Prentice, E.J, Summers, E.L. | | 登録日 | 2021-06-30 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Serine acetyltransferase from Neisseria gonorrhoeae; structural and biochemical basis of inhibition.
Biochem.J., 479, 2022
|
|
7RK9
 
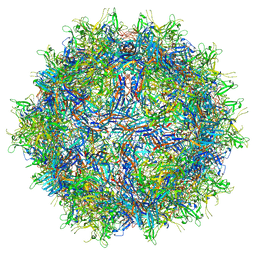 | |
2GAF
 
 | | Crystal Structure of the Vaccinia Polyadenylate Polymerase Heterodimer (apo form) | | 分子名称: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit | | 著者 | Moure, C.M, Bowman, B.R, Gershon, P.D, Quiocho, F.A. | | 登録日 | 2006-03-08 | | 公開日 | 2006-05-16 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity.
Mol.Cell, 22, 2006
|
|
4FHG
 
 | | Spore photoproduct lyase C140S mutant | | 分子名称: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | 著者 | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | 登録日 | 2012-06-06 | | 公開日 | 2012-07-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
2GDG
 
 | |
4FLO
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | 著者 | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | 登録日 | 2012-06-15 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
2GB2
 
 | | The P52G mutant of amicyanin in the Cu(II) state. | | 分子名称: | Amicyanin, COPPER (II) ION | | 著者 | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | 登録日 | 2006-03-09 | | 公開日 | 2006-08-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
4FO1
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lincosamide resistance protein | | 著者 | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-06-20 | | 公開日 | 2012-07-04 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo
TO BE PUBLISHED
|
|
4F9C
 
 | | Human CDC7 kinase in complex with DBF4 and XL413 | | 分子名称: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, Cell division cycle 7-related protein kinase, Protein DBF4 homolog A, ... | | 著者 | Hughes, S, Cherepanov, P. | | 登録日 | 2012-05-18 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of human CDC7 kinase in complex with its activator DBF4.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1MSS
 
 | |
4FD9
 
 | |
1R4W
 
 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | 分子名称: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | 著者 | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | 登録日 | 2003-10-08 | | 公開日 | 2004-02-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
4FE7
 
 | | structure of xylose-binding transcription activator xylR | | 分子名称: | Xylose operon regulatory protein, alpha-D-xylopyranose | | 著者 | Ni, L, Schumacher, M.A. | | 登録日 | 2012-05-29 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain.
Nucleic Acids Res., 41, 2013
|
|
