8BU9
 
 | | Structure of DDB1 bound to roscovitine-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUK
 
 | | Structure of DDB1 bound to DS08-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-(naphthalen-2-ylmethylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
2BKZ
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-404611 | | 分子名称: | 1-[4-(AMINOSULFONYL)PHENYL]-1,6-DIHYDROPYRAZOLO[3,4-E]INDAZOLE-3-CARBOXAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | 著者 | DAlessio, R, Bargiottia, A, Metz, S, Brasca, M.G, Cameron, A, Ermoli, A, Marsiglio, A, Polucci, P, Roletto, F, Tibolla, M, Vazquez, M.L, Vulpetti, A, Pevarello, P. | | 登録日 | 2005-02-23 | | 公開日 | 2006-03-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Benzodipyrazoles: A New Class of Potent Cdk2 Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8BUP
 
 | | Structure of DDB1 bound to DS30-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[3-(3-methylphenyl)propylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUB
 
 | | Structure of DDB1 bound to dCeMM4-engaged CDK12-cyclin K | | 分子名称: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU3
 
 | | Structure of DDB1 bound to DS19-engaged CDK12-cyclin K | | 分子名称: | 1,2-ETHANEDIOL, 2-morpholin-4-yl-9-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]purin-6-amine, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUL
 
 | | Structure of DDB1 bound to DS11-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-(3-phenylpropylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUQ
 
 | | Structure of DDB1 bound to DS43-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[1-(3-chlorophenyl)pyrazol-3-yl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU5
 
 | | Structure of DDB1 bound to SR-4835-engaged CDK12-cyclin K | | 分子名称: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.134 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUS
 
 | | Structure of DDB1 bound to DS59-engaged CDK12-cyclin K | | 分子名称: | 1,3-dimethyl-5-[[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)methylamino]purin-2-yl]amino]methyl]pyrazole-4-sulfonamide, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.26 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
2BHH
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 4-HYDROXYPIPERINDINESULFONYL-INDIRUBINE | | 分子名称: | (2E,3S)-3-HYDROXY-5'-[(4-HYDROXYPIPERIDIN-1-YL)SULFONYL]-3-METHYL-1,3-DIHYDRO-2,3'-BIINDOL-2'(1'H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | 登録日 | 2005-01-11 | | 公開日 | 2005-03-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
8BUH
 
 | | Structure of DDB1 bound to WX3-engaged CDK12-cyclin K | | 分子名称: | 6-[[[2-[[(2~{R})-1-oxidanylbutan-2-yl]amino]-9-propan-2-yl-purin-6-yl]amino]methyl]-3-pyridin-2-yl-1~{H}-pyridin-2-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.79 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUC
 
 | | Structure of DDB1 bound to dCeMM3-engaged CDK12-cyclin K | | 分子名称: | 2-(1~{H}-benzimidazol-2-ylsulfanyl)-~{N}-(5-chloranylpyridin-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUT
 
 | | Structure of DDB1 bound to DS61-engaged CDK12-cyclin K | | 分子名称: | 2-[[6-[[4-(2-hydroxyethyloxy)phenyl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUD
 
 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUE
 
 | | Structure of DDB1 bound to Z11-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUG
 
 | | Structure of DDB1 bound to HQ461-engaged CDK12-cyclin K | | 分子名称: | 2-[2-[(6-methylpyridin-2-yl)amino]-1,3-thiazol-4-yl]-~{N}-(5-methyl-1,3-thiazol-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.53 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
2C68
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | 分子名称: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDROPYRAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | 登録日 | 2005-11-08 | | 公開日 | 2005-12-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZZ3
 
 | | Crystal structure of a HDAC-like protein with CypX bound | | 分子名称: | 3-CYCLOPENTYL-N-HYDROXYPROPANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | 著者 | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | 登録日 | 2005-06-13 | | 公開日 | 2005-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
5J9T
 
 | | Crystal structure of the NuA4 core complex | | 分子名称: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | 著者 | Chen, Z.C, Xu, P. | | 登録日 | 2016-04-11 | | 公開日 | 2016-10-26 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
3JVM
 
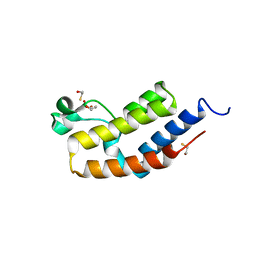 | | Crystal structure of bromodomain 2 of mouse Brd4 | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | 著者 | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | 登録日 | 2009-09-17 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
4LV9
 
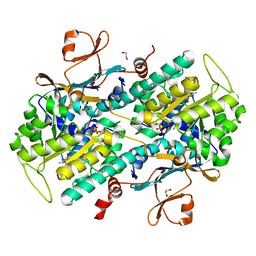 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, 7-chloro-3-methyl-2H-1,2,4-benzothiadiazine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | 登録日 | 2013-07-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.807 Å) | | 主引用文献 | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7D8X
 
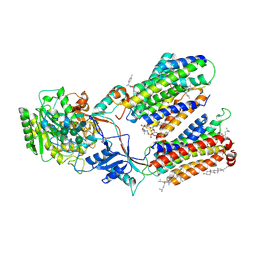 | | CryoEM structure of human gamma-secretase in complex with E2012 and L685458 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(1S)-1-(4-fluorophenyl)ethyl]-3-[[3-methoxy-4-(4-methylimidazol-1-yl)phenyl]methylidene]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | 登録日 | 2020-10-11 | | 公開日 | 2021-01-27 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7NR4
 
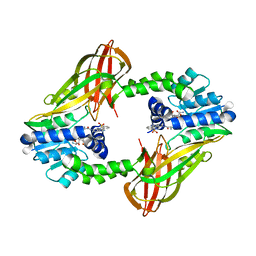 | | X-RAY STRUCTURE OF PRMT6 IN COMPLEX WITH indazole type inhibitor | | 分子名称: | (2~{S})-2-azanyl-~{N}-[3-[3-(dimethylsulfamoyl)phenyl]-2~{H}-indazol-5-yl]propanamide, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Steuber, H. | | 登録日 | 2021-03-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Rational Design and Synthesis of Selective PRMT4 Inhibitors: A New Chemotype for Development of Cancer Therapeutics*.
Chemmedchem, 16, 2021
|
|
