8RDW
 
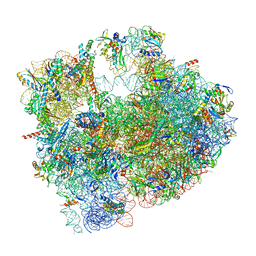 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3). | | 分子名称: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | 著者 | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | 登録日 | 2023-12-08 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8RD8
 
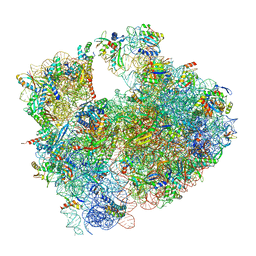 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1). | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S30, ... | | 著者 | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | 登録日 | 2023-12-07 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
6GFJ
 
 | |
4YS9
 
 | |
8J2P
 
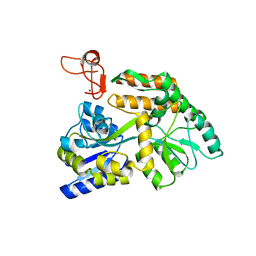 | | Crystal structure of PML B-box2 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Zhou, C, Zang, N, Zhang, J. | | 登録日 | 2023-04-15 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
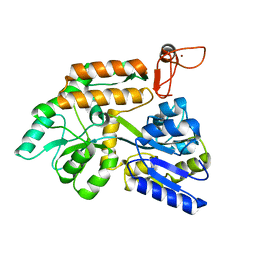 | | Crystal structure of PML B-box2 mutant | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Zhou, C, Zang, N, Zhang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
7WCC
 
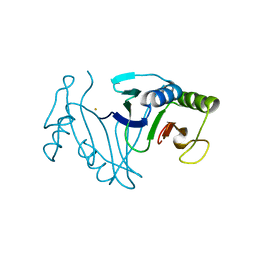 | | Oxidase ChaP-D49L/Q91C mutant | | 分子名称: | ChaP, FE (III) ION | | 著者 | Sun, M.X, Zheng, W, Wang, Y.S, Zhu, J.P, Tan, R.X. | | 登録日 | 2021-12-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Oxidase ChaP-D49L/Q91C mutant
To Be Published
|
|
5KY9
 
 | |
8QSO
 
 | | Crystal structure of human Mcl-1 in complex with compound 1 | | 分子名称: | (13S,16R,19S)-16-benzyl-43-ethoxy-N-methyl-7,11,14,17-tetraoxo-13-phenyl-5-oxa-2,8,12,15,18-pentaaza-1(1,4),4(1,2)-dibenzena-9(1,4)-cyclohexanacycloicosaphane-19-carboxamide, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Hekking, K.F.W, Gremmen, S, Maroto, S, Keefe, A.D, Zhang, Y. | | 登録日 | 2023-10-10 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Development of Potent Mcl-1 Inhibitors: Structural Investigations on Macrocycles Originating from a DNA-Encoded Chemical Library Screen.
J.Med.Chem., 67, 2024
|
|
2Y5S
 
 | | Crystal structure of Burkholderia cenocepacia dihydropteroate synthase complexed with 7,8-dihydropteroate. | | 分子名称: | 1,2-ETHANEDIOL, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | 著者 | Morgan, R.E, Batot, G.O, Dement, J.M, Rao, V.A, Eadsforth, T.C, Hunter, W.N. | | 登録日 | 2011-01-17 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structures of Burkholderia Cenocepacia Dihydropteroate Synthase in the Apo-Form and Complexed with the Product 7,8-Dihydropteroate.
Bmc Struct.Biol., 11, 2011
|
|
5KY0
 
 | |
8JBG
 
 | | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq (G324), ... | | 著者 | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | 登録日 | 2023-05-08 | | 公開日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
8JBH
 
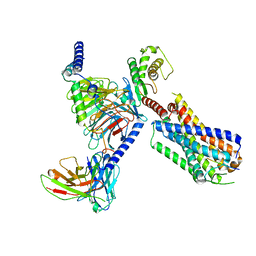 | | Substance P bound to active human neurokinin 3 receptor in complex with Gq | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein Gq subunit alpha (G324), ... | | 著者 | Sun, W.J, Yang, F, Zhang, H.H, Yuan, Q.N, Yin, W.C, Shi, P, Eric, X, Tian, C.L. | | 登録日 | 2023-05-08 | | 公開日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists.
Cell Discov, 9, 2023
|
|
6HDC
 
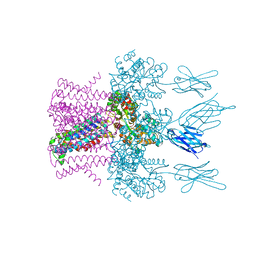 | | Crystal structure of the potassium channel MtTMEM175 T38A variant in complex with a Nanobody-MBP fusion protein | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | 著者 | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | 登録日 | 2018-08-17 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDB
 
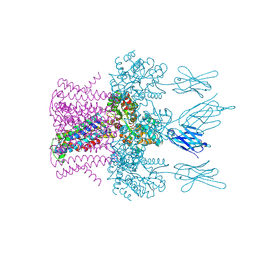 | | Crystal structure of the potassium channel MtTMEM175 with zinc | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | 著者 | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | 登録日 | 2018-08-17 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
8IZJ
 
 | |
6QYL
 
 | | Structure of MBP-Mcl-1 in complex with compound 8a | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-09 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6HGP
 
 | |
1LY8
 
 | | The crystal structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability and insight into modelling of protein structures | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | 著者 | Houborg, K, Harris, P, Poulsen, J.-C.N, Svendsen, A, Schneider, P, Larsen, S. | | 登録日 | 2002-06-07 | | 公開日 | 2002-06-14 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6HD9
 
 | | Crystal structure of the potassium channel MtTMEM175 with rubidium | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, RUBIDIUM ION, ... | | 著者 | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | 登録日 | 2018-08-17 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6QUG
 
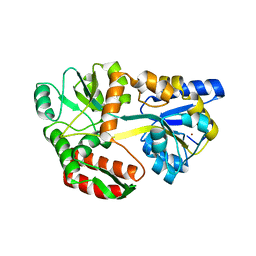 | | GHK tagged MBP-Nup98(1-29) | | 分子名称: | COPPER (II) ION, Maltodextrin-binding protein,Nucleoporin, putative, ... | | 著者 | Huyton, T, Gorlich, D. | | 登録日 | 2019-02-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1MH3
 
 | |
8SPU
 
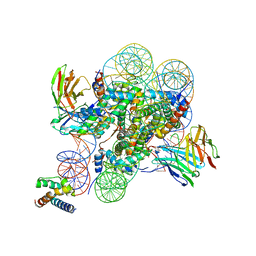 | | Structure of ESRRB nucleosome bound OCT4 at site c | | 分子名称: | DNA (168-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-03 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
8SPS
 
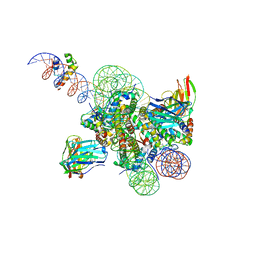 | | High resolution structure of ESRRB nucleosome bound OCT4 at site a and site b | | 分子名称: | DNA (168-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-03 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
8T2I
 
 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | 分子名称: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | 著者 | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | 登録日 | 2023-06-06 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (10.4 Å) | | 主引用文献 | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
