8A6H
 
 | | Small molecule stabilizer (compound 7) for C-RAF and 14-3-3 | | 分子名称: | 14-3-3 protein sigma, 3-[2-(dimethylamino)ethyldisulfanyl]-1-[4-oxidanyl-4-[3-(trifluoromethyl)phenyl]piperidin-1-yl]propan-1-one, CHLORIDE ION, ... | | 著者 | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | 登録日 | 2022-06-17 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
4NUD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS436 inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | 著者 | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | 登録日 | 2013-12-03 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
8A65
 
 | | Small molecule stabilizer (compound 3) for FOXO1 and 14-3-3 | | 分子名称: | (3~{S})-1-[2-azanyl-3,5-bis(chloranyl)phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | 著者 | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | 登録日 | 2022-06-16 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A68
 
 | | Small molecule stabilizer (compound 5) for C-RAF(pS259) and 14-3-3 | | 分子名称: | 14-3-3 protein sigma, MAGNESIUM ION, RAF proto-oncogene serine/threonine-protein kinase, ... | | 著者 | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | 登録日 | 2022-06-16 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A6F
 
 | | Small molecule stabilizer (compound 8) for C-RAF and 14-3-3 | | 分子名称: | 1-[4-[4-chloranyl-3-(trifluoromethyl)phenyl]-4-oxidanyl-piperidin-1-yl]-3-[2-(dimethylamino)ethyldisulfanyl]propan-1-one, 14-3-3 protein sigma, CHLORIDE ION, ... | | 著者 | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | 登録日 | 2022-06-17 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
1ZZ1
 
 | | Crystal structure of a HDAC-like protein with SAHA bound | | 分子名称: | Histone deacetylase-like amidohydrolase, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, POTASSIUM ION, ... | | 著者 | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | 登録日 | 2005-06-13 | | 公開日 | 2005-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
7ZW7
 
 | |
2AFG
 
 | |
7LP0
 
 | |
7LRK
 
 | |
7LPK
 
 | |
7LRO
 
 | |
3L30
 
 | | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine | | 分子名称: | 4,14-dihydro-8H-[1,3]dioxolo[4,5-g]isoquino[3,2-a]isoquinoline-9,10-diol, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Naveen, C, Prasanth, G.K, Abhilash, J, Pradeep, M, Ponnuraj, K, Sadasivan, C, Haridas, M. | | 登録日 | 2009-12-16 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine
To be Published
|
|
7ZHF
 
 | |
7ZHK
 
 | |
8AG2
 
 | | Crystal structure of the BPTF bromodomain in complex with BI-7190 | | 分子名称: | 5-[3-methoxy-4-[1-(4-methylpiperazin-1-yl)cyclopropyl]phenyl]-1,3,4-trimethyl-pyridin-2-one, Nucleosome-remodeling factor subunit BPTF | | 著者 | Bader, G, Boettcher, J, Wolkerstorfer, B. | | 登録日 | 2022-07-19 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.025 Å) | | 主引用文献 | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
7M2F
 
 | | CDK2 with compound 14 inhibitor with carboxylate | | 分子名称: | Cyclin-dependent kinase 2, [(1r,4r)-4-{4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-3,6-dihydropyridin-1(2H)-yl}cyclohexyl]acetic acid | | 著者 | Longenecker, K.L, Qiu, W, Korepanova, A, Tong, Y. | | 登録日 | 2021-03-16 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.632 Å) | | 主引用文献 | Balancing Properties with Carboxylates: A Lead Optimization Campaign for Selective and Orally Active CDK9 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8AG0
 
 | | Crystal structure of mutant PRELID3a-TRIAP1 complex - R53E | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3A, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Milara, X, Perez-Dorado, J.I, Matthews, S.J. | | 登録日 | 2022-07-18 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | An intermolecular hydrogen bonded network in the PRELID-TRIAP protein family plays a role in lipid sensing.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
2ATX
 
 | | Crystal Structure of the TC10 GppNHp complex | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, small GTP binding protein TC10 | | 著者 | Hemsath, L, Dvorsky, R, Fiegen, D, Carlier, M.F, Ahmadian, M.R. | | 登録日 | 2005-08-26 | | 公開日 | 2005-09-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins
Mol.Cell, 20, 2005
|
|
3L5X
 
 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | 著者 | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | 登録日 | 2009-12-22 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
2AJ4
 
 | | Crystal structure of Saccharomyces cerevisiae Galactokinase in complex with galactose and Mg:AMPPNP | | 分子名称: | CHLORIDE ION, Galactokinase, MAGNESIUM ION, ... | | 著者 | Thoden, J.B, Sellick, C.A, Timson, D.J, Reece, R.J, Holden, H.M. | | 登録日 | 2005-08-01 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer
J.Biol.Chem., 280, 2005
|
|
2ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | 分子名称: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | 著者 | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | 登録日 | 1999-01-20 | | 公開日 | 2000-01-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
8AFN
 
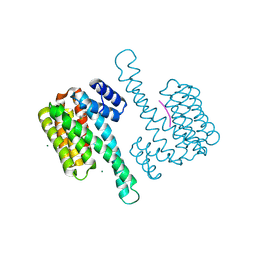 | | Small molecule stabilizer (compound 1) for ERalpha and 14-3-3 | | 分子名称: | 1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-4-carboxamide, 14-3-3 protein sigma, Estrogen receptor, ... | | 著者 | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | 登録日 | 2022-07-18 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | 分子名称: | Regulatory protein NPR1, ZINC ION | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
3FVI
 
 | |
