6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6GVS
 
 | | Engineered glycolyl-CoA reductase comprising 8 mutations with bound NADP+ | | 分子名称: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION | | 著者 | Zarzycki, J, Trudeau, D, Scheffen, M, Erb, T.J, Tawfik, D.S. | | 登録日 | 2018-06-21 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.579 Å) | | 主引用文献 | Design and in vitro realization of carbon-conserving photorespiration.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1XRK
 
 | | Crystal structure of a mutant bleomycin binding protein from Streptoalloteichus hindustanus displaying increased thermostability | | 分子名称: | BLEOMYCIN A2, Bleomycin resistance protein, SULFATE ION | | 著者 | Brouns, S.J.J, Wu, H, Akerboom, J, Turnbull, A.P, de Vos, W.M, Van der Oost, J. | | 登録日 | 2004-10-15 | | 公開日 | 2005-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Engineering a selectable marker for hyperthermophiles
J.Biol.Chem., 280, 2005
|
|
7XUP
 
 | | Crystal structure of TPe3.0 | | 分子名称: | Transcriptional regulator, PadR-like family | | 著者 | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | 登録日 | 2022-05-19 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
1FXY
 
 | | COAGULATION FACTOR XA-TRYPSIN CHIMERA INHIBITED WITH D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | 分子名称: | COAGULATION FACTOR XA-TRYPSIN CHIMERA, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide | | 著者 | Hopfner, K.P, Kopetzki, E, Kresse, G.-B, Huber, R, Bode, W, Engh, R.A. | | 登録日 | 1998-04-22 | | 公開日 | 1998-06-17 | | 最終更新日 | 2013-02-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | New enzyme lineages by subdomain shuffling.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4HVY
 
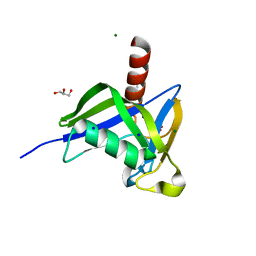 | | A thermostable variant of human NUDT18 NUDIX domain obtained by Hot Colony Filtration | | 分子名称: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate-linked moiety X motif 18, ... | | 著者 | Asial, I, Cheng, Y.X, Engman, H, Wu, B, Dollhopf, M, Nordlund, P, Cornvik, T. | | 登録日 | 2012-11-07 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell.
Nat Commun, 4, 2013
|
|
3LB9
 
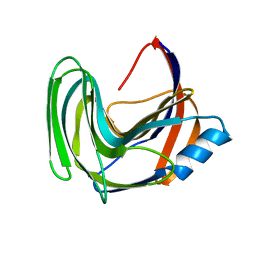 | | Crystal structure of the B. circulans cpA123 circular permutant | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
3LTM
 
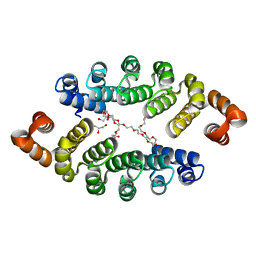 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | 分子名称: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | 著者 | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | 登録日 | 2010-02-16 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
8XEG
 
 | |
3LTD
 
 | | X-ray structure of a non-biological ATP binding protein determined at 2.8 A by multi-wavelength anomalous dispersion | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | 著者 | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | 登録日 | 2010-02-15 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
3LTJ
 
 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | 分子名称: | AlphaRep-4 | | 著者 | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | 登録日 | 2010-02-16 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
7JU0
 
 | |
2ZM8
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | 登録日 | 2008-04-14 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
3BVG
 
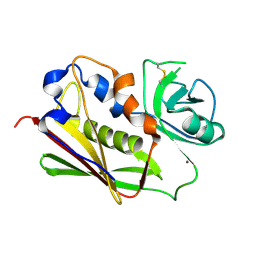 | |
7LTM
 
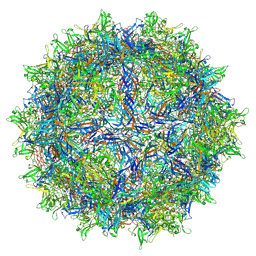 | | Hum8 capsid | | 分子名称: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | 著者 | Mietzsch, M, Agbandje-McKenna, M. | | 登録日 | 2021-02-19 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Receptor Switching in Newly Evolved Adeno-associated Viruses.
J.Virol., 95, 2021
|
|
8UKE
 
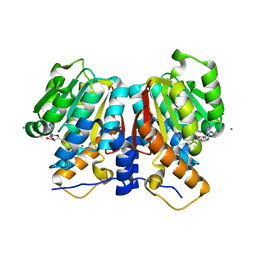 | |
2ZLY
 
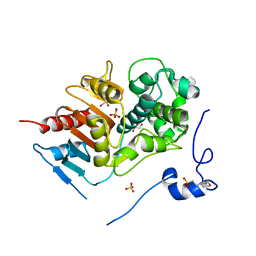 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM9
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) with Substrate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | 登録日 | 2008-04-14 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
7MKV
 
 | |
2ZM2
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
6IZC
 
 | |
6J1Q
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - RS | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Cen, Y.X, Zhou, J.H, Wu, Q. | | 登録日 | 2018-12-29 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
4OS7
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK607 (bicyclic) | | 分子名称: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Chen, S, Pojer, F, Heinis, C. | | 登録日 | 2014-02-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
6IYL
 
 | | The structure of EntE with 3-cyanobenzoyl adenylate analog | | 分子名称: | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex, 5'-O-[(3-cyanobenzene-1-carbonyl)sulfamoyl]adenosine | | 著者 | Miyanaga, A, Ishikawa, F. | | 登録日 | 2018-12-17 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | An Engineered Aryl Acid Adenylation Domain with an Enlarged Substrate Binding Pocket.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6IZD
 
 | |
