5BVA
 
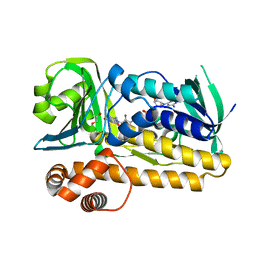 | | Structure of flavin-dependent brominase Bmp2 | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase | | 著者 | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | 登録日 | 2015-06-04 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.873 Å) | | 主引用文献 | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BUK
 
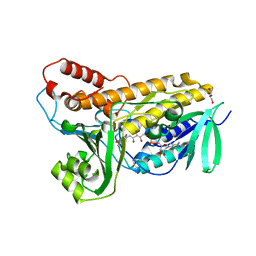 | | Structure of flavin-dependent chlorinase Mpy16 | | 分子名称: | FADH2-dependent halogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | 登録日 | 2015-06-03 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2XYO
 
 | | Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | 登録日 | 2010-11-18 | | 公開日 | 2011-03-23 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
2XDO
 
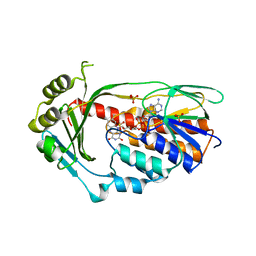 | | Structure of the Tetracycline degrading Monooxygenase TetX2 from Bacteroides thetaiotaomicron | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN | | 著者 | Volkers, G, Palm, G.J, Wright, G.D, Hinrichs, W. | | 登録日 | 2010-05-05 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
8TWF
 
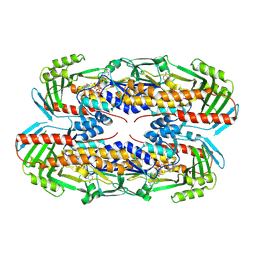 | |
8TWG
 
 | |
8UIQ
 
 | | H47Q NicC with 2-mercaptopyridine ligand | | 分子名称: | 2-PYRIDINETHIOL, 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Hicks, K.A, Perry, K, Turlington, Z.R, Vaz Ferreira de Macedo, S. | | 登録日 | 2023-10-10 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
8UIV
 
 | | H47Q NicC with bound FAD | | 分子名称: | 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Hicks, K.A, Perry, K. | | 登録日 | 2023-10-10 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.511 Å) | | 主引用文献 | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
3ALL
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant Y270A | | 分子名称: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | 登録日 | 2010-08-04 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be published
|
|
2Y6Q
 
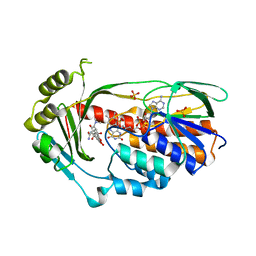 | | Structure of the TetX monooxygenase in complex with the substrate 7- Iodtetracycline | | 分子名称: | 7-IODOTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | 登録日 | 2011-01-25 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
2Y6R
 
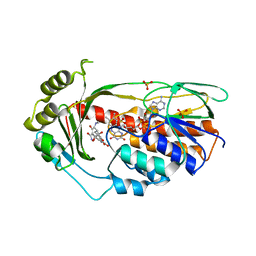 | | Structure of the TetX monooxygenase in complex with the substrate 7- chlortetracycline | | 分子名称: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | 登録日 | 2011-01-25 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
8WVB
 
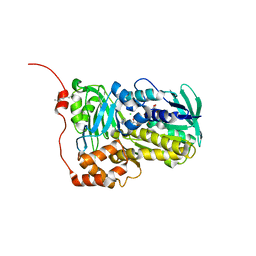 | | Crystal structure of Lsd18 mutant S195M | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | 著者 | Liu, N, Xiao, H.L, Chen, X. | | 登録日 | 2023-10-23 | | 公開日 | 2023-12-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
8WVF
 
 | |
2PHH
 
 | |
3V3O
 
 | |
3V3N
 
 | | Crystal structure of TetX2 T280A: an adaptive mutant in complex with minocycline | | 分子名称: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Walkiewicz, K, Shamoo, Y. | | 登録日 | 2011-12-13 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Crystal structure of TetX2 T280A: an adaptive mutant in complex with minocycline
To be Published
|
|
3C96
 
 | | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR240 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | 著者 | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-02-15 | | 公開日 | 2008-02-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa.
To be Published
|
|
4A6N
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH TIGECYCLINE | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | 登録日 | 2011-11-07 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ALJ
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, reduced form | | 分子名称: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | 登録日 | 2010-08-04 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
3ALM
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant C294A | | 分子名称: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | 登録日 | 2010-08-04 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4A99
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH MINOCYCLINE | | 分子名称: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | 登録日 | 2011-11-25 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BJZ
 
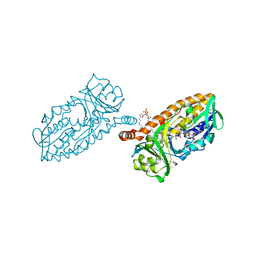 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Native data | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | 著者 | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | 登録日 | 2013-04-21 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4BJY
 
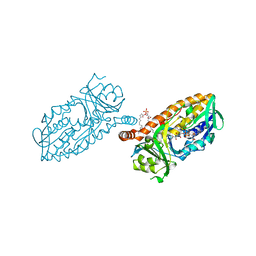 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Platinum derivative | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | 著者 | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | 登録日 | 2013-04-21 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
5TUM
 
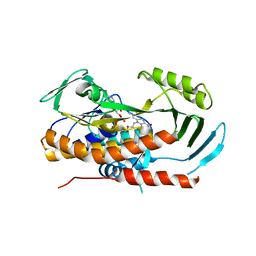 | |
5TUF
 
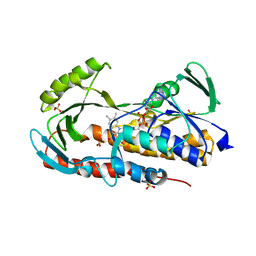 | | Crystal structure of tetracycline destructase Tet(50) in complex with anhydrotetracycline | | 分子名称: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Park, J, Tolia, N.H. | | 登録日 | 2016-11-06 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
