8PN8
 
 | | Engineered glycolyl-CoA carboxylase (L100N variant) with bound CoA | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | 著者 | Zarzycki, J, Marchal, D.G, Schulz, L, Prinz, S, Erb, T.J. | | 登録日 | 2023-06-30 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.31 Å) | | 主引用文献 | Machine Learning-Supported Enzyme Engineering toward Improved CO 2 -Fixation of Glycolyl-CoA Carboxylase.
Acs Synth Biol, 12, 2023
|
|
2ORI
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/) | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Counago, R, Shamoo, Y, Wilson, C.J, Wu, G, Myers, J. | | 登録日 | 2007-02-02 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/)
To be Published
|
|
2OO7
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R) | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Counago, R, Wilson, C.J, Wu, G, Myers, J.C, Wittung-Stafshede, P, Shamoo, Y. | | 登録日 | 2007-01-25 | | 公開日 | 2008-02-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R)
To be Published
|
|
2OSB
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/) | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Myers, J, Counago, R, Wilson, C.J, Wu, G, Shamoo, Y. | | 登録日 | 2007-02-05 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/)
To be Published
|
|
2QAJ
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q199R/G213E) | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Counago, R, Wilson, C.J, Myers, J, Wu, G, Shamoo, Y. | | 登録日 | 2007-06-15 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q199R/G213E)
To be Published
|
|
4ZV6
 
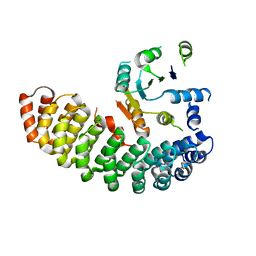 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | 分子名称: | AlphaRep-7, Octarellin V.1 | | 著者 | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | 登録日 | 2015-05-18 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6GVS
 
 | | Engineered glycolyl-CoA reductase comprising 8 mutations with bound NADP+ | | 分子名称: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION | | 著者 | Zarzycki, J, Trudeau, D, Scheffen, M, Erb, T.J, Tawfik, D.S. | | 登録日 | 2018-06-21 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.579 Å) | | 主引用文献 | Design and in vitro realization of carbon-conserving photorespiration.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7XUP
 
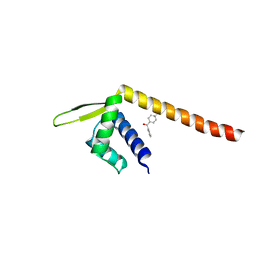 | | Crystal structure of TPe3.0 | | 分子名称: | Transcriptional regulator, PadR-like family | | 著者 | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | 登録日 | 2022-05-19 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7XUQ
 
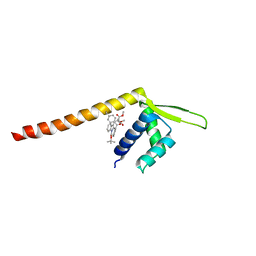 | | Crystal structure of Tpe3.0 complexed with N-Boc-3-alkenylindole | | 分子名称: | Transcriptional regulator, PadR-like family, dimethyl 2-[[2-methyl-1-[(2-methylpropan-2-yl)oxycarbonyl]indol-3-yl]methyl]-2-prop-2-enyl-propanedioate | | 著者 | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | 登録日 | 2022-05-19 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
2ZM8
 
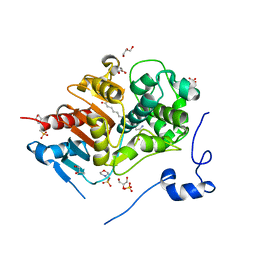 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | 登録日 | 2008-04-14 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
5BOP
 
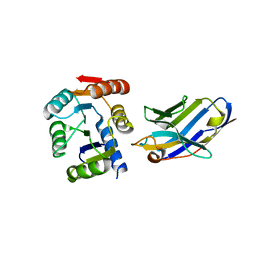 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | 分子名称: | Nanobody, Octarellin V.1 | | 著者 | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | 登録日 | 2015-05-27 | | 公開日 | 2016-05-25 | | 最終更新日 | 2016-06-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
7DNA
 
 | |
2RKX
 
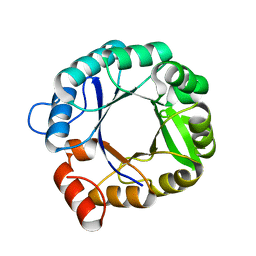 | | The 3D structure of chain D, cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | 分子名称: | Cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | 著者 | Tawfik, D, Khersonsky, O, Albeck, S, Dym, O, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2007-10-18 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Kemp elimination catalysts by computational enzyme design.
Nature, 453, 2008
|
|
7DMX
 
 | |
7DNB
 
 | |
7BDN
 
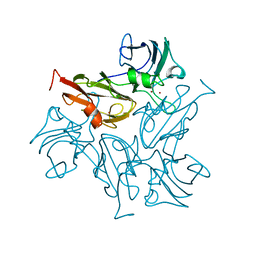 | | Structure of the Streptomyces coelicolor small laccase - cubic crystal form | | 分子名称: | COPPER (II) ION, Putative copper oxidase | | 著者 | Zovo, K, Majumdar, S, Lukk, T. | | 登録日 | 2020-12-22 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Substitution of the Methionine Axial Ligand of the T1 Copper for the Fungal-like Phenylalanine Ligand (M298F) Causes Local Structural Perturbations that Lead to Thermal Instability and Reduced Catalytic Efficiency of the Small Laccase from Streptomyces coelicolor A3(2).
Acs Omega, 7, 2022
|
|
7BFM
 
 | |
2ZM2
 
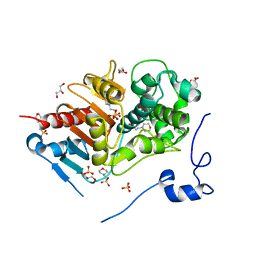 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
6BRA
 
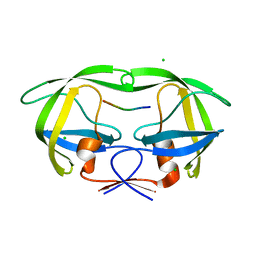 | |
7EYP
 
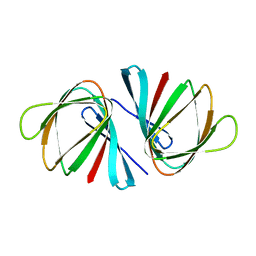 | |
7EYK
 
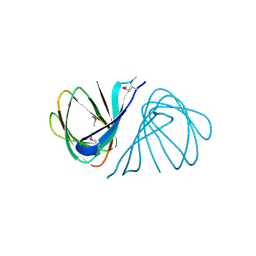 | |
7EYJ
 
 | | Crystal structure of Escherichia coli ppnP | | 分子名称: | Pyrimidine/purine nucleoside phosphorylase, SULFATE ION | | 著者 | Wen, Y, Wu, B.X. | | 登録日 | 2021-05-31 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYL
 
 | | Crystal structure of Salmonella enterica ppnP | | 分子名称: | Pyrimidine/purine nucleoside phosphorylase | | 著者 | Wen, Y, Wu, B.X. | | 登録日 | 2021-05-31 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
7EYM
 
 | | Crystal structure of Vibrio cholerae ppnP | | 分子名称: | Pyrimidine/purine nucleoside phosphorylase | | 著者 | Wen, Y, Wu, B.X. | | 登録日 | 2021-05-31 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Crystal structures of a new class of pyrimidine/purine nucleoside phosphorylase revealed a Cupin fold.
Proteins, 90, 2022
|
|
2ZLY
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
