5JMG
 
 | | X-ray structure of the complex between bovine pancreatic ribonuclease and pentachlorocarbonyliridate(III) (4 days of soaking) | | 分子名称: | CARBON MONOXIDE, CHLORIDE ION, IRIDIUM ION, ... | | 著者 | Caterino, M, Petruk, A.A, Vergara, A, Ferraro, G, Merlino, A. | | 登録日 | 2016-04-29 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Mapping the protein-binding sites for iridium(iii)-based CO-releasing molecules.
Dalton Trans, 45, 2016
|
|
1AE5
 
 | | HUMAN HEPARIN BINDING PROTEIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEPARIN BINDING PROTEIN | | 著者 | Iversen, L.F, Kastrup, J.S, Bjorn, S.E, Rasmussen, P.B, Wiberg, F.C, Flodgaard, H.J, Larsen, I.K. | | 登録日 | 1997-03-05 | | 公開日 | 1998-03-11 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of HBP, a multifunctional protein with a serine proteinase fold.
Nat.Struct.Biol., 4, 1997
|
|
1LNX
 
 | | Crystal structure of the P.aerophilum SmAP1 heptamer in a new crystal form (C2221) | | 分子名称: | ACETIC ACID, GLYCEROL, URIDINE, ... | | 著者 | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | 登録日 | 2002-05-04 | | 公開日 | 2003-03-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
5F9A
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain P436 in complex with blood group H Lewis b hexasaccharide | | 分子名称: | Adhesin binding fucosylated histo-blood group antigen,Adhesin,Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER19, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | 登録日 | 2015-12-09 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
1LQ7
 
 | | De Novo Designed Protein Model of Radical Enzymes | | 分子名称: | Alpha3W | | 著者 | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | 登録日 | 2002-05-09 | | 公開日 | 2002-06-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
1GMY
 
 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | 分子名称: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | 著者 | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | 登録日 | 2001-09-25 | | 公開日 | 2002-09-19 | | 最終更新日 | 2017-07-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
2C36
 
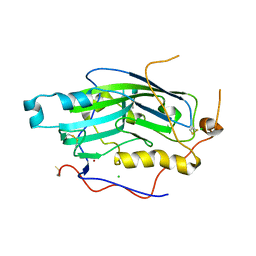 | | Structure of unliganded HSV gD reveals a mechanism for receptor- mediated activation of virus entry | | 分子名称: | CHLORIDE ION, GLYCOPROTEIN D HSV-1, ZINC ION, ... | | 著者 | Krummenacher, C, Supekar, V.M, Whitbeck, J.C, Lazear, E, Connolly, S.A, Eisenberg, R.J, Cohen, G.H, Wiley, D.C, Carfi, A. | | 登録日 | 2005-10-04 | | 公開日 | 2005-11-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry.
EMBO J., 24, 2005
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | 分子名称: | IMMUNOGLOBULIN G BINDING PROTEIN G | | 著者 | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | 登録日 | 2001-08-02 | | 公開日 | 2001-08-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJS
 
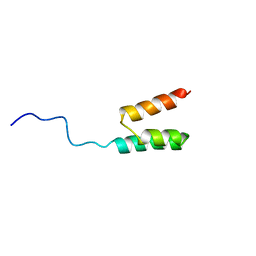 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | 分子名称: | IMMUNOGLOBULIN G BINDING PROTEIN G | | 著者 | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | 登録日 | 2001-08-02 | | 公開日 | 2001-08-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
5FMF
 
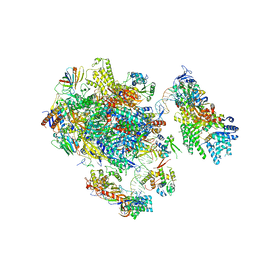 | | the P-lobe of RNA polymerase II pre-initiation complex | | 分子名称: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | 著者 | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | 登録日 | 2015-11-03 | | 公開日 | 2015-11-25 | | 最終更新日 | 2017-07-12 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KYY
 
 | |
5FOJ
 
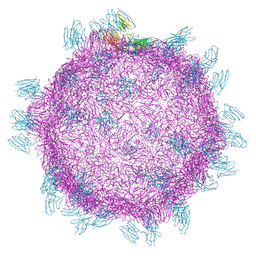 | | Cryo electron microscopy structure of Grapevine Fanleaf Virus complex with Nanobody | | 分子名称: | Nanobody, RNA2 polyprotein | | 著者 | Orlov, I, Hemmer, C, Ackerer, L, Lorber, B, Ghannam, A, Poignavent, V, Hleibieh, K, Sauter, C, Schmitt-Keichinger, C, Belval, L, Hily, J.M, Marmonier, A, Komar, V, Gersch, S, Schellenberger, P, Bron, P, Vigne, E, Muyldermans, S, Lemaire, O, Demangeat, G, Ritzenthaler, C, Klaholz, B.P. | | 登録日 | 2015-11-22 | | 公開日 | 2016-01-20 | | 最終更新日 | 2021-08-11 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of nanobody recognition of grapevine fanleaf virus and of virus resistance loss.
Proc.Natl.Acad.Sci.USA, 2020
|
|
5KYX
 
 | |
3B4T
 
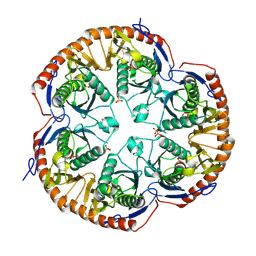 | | Crystal structure of Mycobacterium tuberculosis RNase PH, the Mycobacterium tuberculosis Structural Genomics Consortium target Rv1340 | | 分子名称: | PHOSPHATE ION, Ribonuclease PH | | 著者 | Antczak, A.J, Berger, J.M, Lekin, T, Segelke, B.W, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB), TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2007-10-24 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 2.1 A Crystal structure of RNase PH from Mycobacterium tuberculosis.
To be Published
|
|
7NWF
 
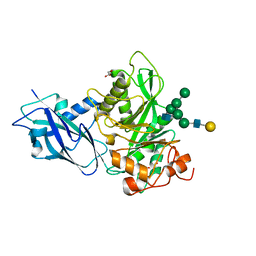 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with hybrid-type glycan (GalGlcNAcMan5GlcNAc) product | | 分子名称: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Trastoy, B, Du, J.J, Garcia-Alija, M, Sundberg, E.J, Guerin, M.E. | | 登録日 | 2021-03-16 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | GH18 endo-beta-N-acetylglucosaminidases use distinct mechanisms to process hybrid-type N-linked glycans.
J.Biol.Chem., 297, 2021
|
|
1GPC
 
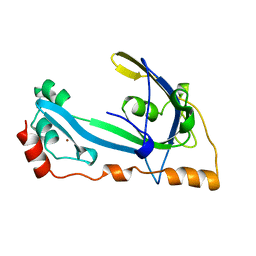 | | CORE GP32, DNA-BINDING PROTEIN | | 分子名称: | PROTEIN (CORE GP32), ZINC ION | | 著者 | Shamoo, Y, Friedman, A.M, Parsons, M.R, Konigsberg, W.H, Steitz, T.A. | | 登録日 | 1995-06-01 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA.
Nature, 376, 1995
|
|
2E2I
 
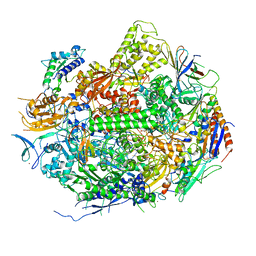 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dGTP | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', ... | | 著者 | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | 登録日 | 2006-11-14 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
7P1T
 
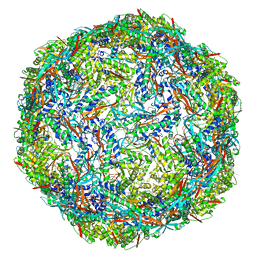 | |
5J07
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P1/2 | | 分子名称: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-27 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J0C
 
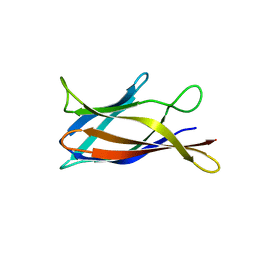 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P2/3 | | 分子名称: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | 著者 | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | 登録日 | 2016-03-28 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
7NVH
 
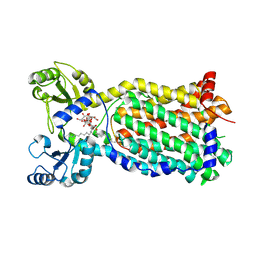 | | Cryo-EM structure of the mycolic acid transporter MmpL3 from M. tuberculosis | | 分子名称: | Lauryl Maltose Neopentyl Glycol, Trehalose monomycolate exporter MmpL3 | | 著者 | Adams, O, Deme, J.C, Parker, J.L, Lea, S.M, Newstead, S. | | 登録日 | 2021-03-15 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: An emergent therapeutic target.
Structure, 29, 2021
|
|
1ALQ
 
 | |
7O9S
 
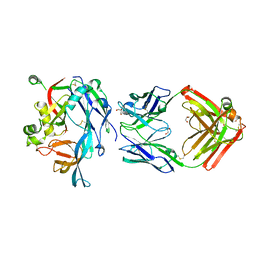 | | Hantaan virus Gn in complex with Fab nnHTN-Gn2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, Fab nnHTN-Gn2 Heavy chain, ... | | 著者 | Rissanen, I, Bowden, T.A, Huiskonen, J.T, Stass, R. | | 登録日 | 2021-04-16 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis for a Neutralizing Antibody Response Elicited by a Recombinant Hantaan Virus Gn Immunogen.
Mbio, 12, 2021
|
|
1HAR
 
 | |
7NRH
 
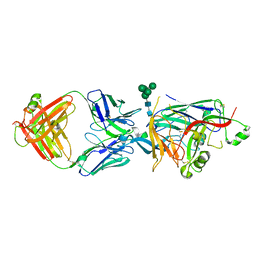 | | Hantaan virus glycoprotein (Gn) in complex with Fab fragment HTN-Gn1. | | 分子名称: | Envelope polyprotein, Fab fragment HTN-Gn1 Heavy chain, Fab fragment HTN-Gn1 Light chain, ... | | 著者 | Stass, R, Rissanen, I, Bowden, T.A, Huiskonen, J.T. | | 登録日 | 2021-03-03 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (19 Å) | | 主引用文献 | Structural Basis for a Neutralizing Antibody Response Elicited by a Recombinant Hantaan Virus Gn Immunogen.
Mbio, 12, 2021
|
|
