7VYQ
 
 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | 分子名称: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | 著者 | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2021-11-15 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DMG
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | 分子名称: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-12-03 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | 分子名称: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLD
 
 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | 分子名称: | Carbonyl Reductase, MAGNESIUM ION | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DN1
 
 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | 分子名称: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-12-08 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | 分子名称: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
3UJ0
 
 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor with ligand bound form. | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | 登録日 | 2011-11-07 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
7LQM
 
 | |
3UJ4
 
 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | 分子名称: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | 著者 | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | 登録日 | 2011-11-07 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
7LQN
 
 | |
1ALG
 
 | | SOLUTION STRUCTURE OF AN HGR INHIBITOR, NMR, 10 STRUCTURES | | 分子名称: | P11 | | 著者 | Schott, M.K, Nordhoff, A, Becker, K, Kalbitzer, H.R, Schirmer, R.H. | | 登録日 | 1997-06-03 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Denaturation and reactivation of dimeric human glutathione reductase--an assay for folding inhibitors.
Eur.J.Biochem., 245, 1997
|
|
1BBB
 
 | |
6MJ3
 
 | | CRYSTAL STRUCTURE OF RHESUS MACAQUE (MACACA MULATTA) IGG1 Fc Fragment-Fc-GAMMA RECEPTOR III complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Igg1 Fc, ... | | 著者 | Gohain, N, Tolbert, W.D, Pazgier, M. | | 登録日 | 2018-09-20 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques.
Front Immunol, 13, 2022
|
|
6MJO
 
 | | CRYSTAL STRUCTURE OF RHESUS MACAQUE (MACACA MULATTA) FC-GAMMA RECEPTOR III | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Low affinity immunoglobulin gamma Fc region receptor III, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Van, V, Tolbert, W.D, Pazgier, M. | | 登録日 | 2018-09-21 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques.
Front Immunol, 13, 2022
|
|
1IC1
 
 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | 著者 | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | 登録日 | 1998-03-09 | | 公開日 | 1998-06-17 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1C2Y
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | 分子名称: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PROTEIN (LUMAZINE SYNTHASE) | | 著者 | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | 登録日 | 1999-07-27 | | 公開日 | 2000-07-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly.
Protein Sci., 8, 1999
|
|
1C41
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | 分子名称: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, LUMAZINE SYNTHASE, SULFATE ION | | 著者 | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | 登録日 | 1999-08-03 | | 公開日 | 2000-08-06 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly
Protein Sci., 8, 1999
|
|
7LC0
 
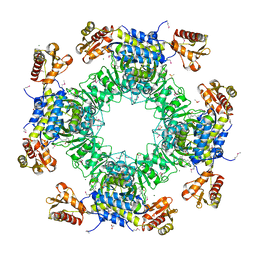 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | 分子名称: | ACETIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Phillips, R.S. | | 登録日 | 2021-01-09 | | 公開日 | 2021-06-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
7LCE
 
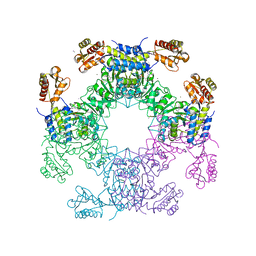 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | 分子名称: | CALCIUM ION, D-glucosaminate-6-phosphate ammonia lyase, DIMETHYL SULFOXIDE | | 著者 | Phillips, R.S. | | 登録日 | 2021-01-10 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
7XU8
 
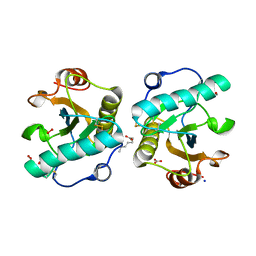 | | Structure of the complex of camel peptidoglycan recognition protein-short (PGRP-S) with heptanoic acid at 2.15 A resolution. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CARBONATE ION, ... | | 著者 | Maurya, A, Ahmad, N, Viswanathan, V, Singh, P.K, Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2022-05-18 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 angstrom resolution.
Int J Biochem Mol Biol, 13, 2022
|
|
3GMV
 
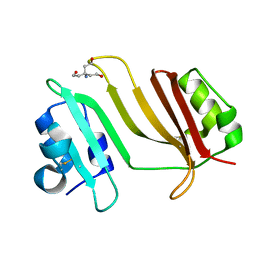 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | 分子名称: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | 著者 | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | 登録日 | 2009-03-15 | | 公開日 | 2009-03-31 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3VAC
 
 | |
3ZQE
 
 | | PrgI-SipD from Salmonella typhimurium in complex with deoxycholate | | 分子名称: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, PROTEIN PRGI, ... | | 著者 | Lunelli, M, Kolbe, M. | | 登録日 | 2011-06-09 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal Structure of Prgi-Sipd: Insight Into a Secretion Competent State of the Type Three Secretion System Needle Tip and its Interaction with Host Ligands
Plos Pathog., 7, 2011
|
|
3N8U
 
 | |
3OYV
 
 | |
