2ROA
 
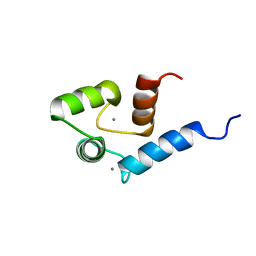 | | Solution structure of calcium bound soybean calmodulin isoform 4 N-terminal domain | | 分子名称: | CALCIUM ION, Calmodulin | | 著者 | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO9
 
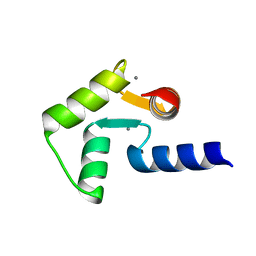 | | Solution structure of calcium bound soybean calmodulin isoform 1 C-terminal domain | | 分子名称: | CALCIUM ION, Calmodulin-2 | | 著者 | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2ROB
 
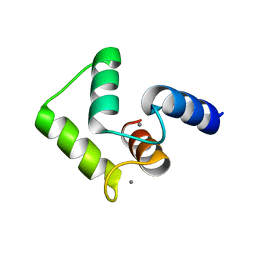 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | 分子名称: | CALCIUM ION, Calmodulin | | 著者 | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO8
 
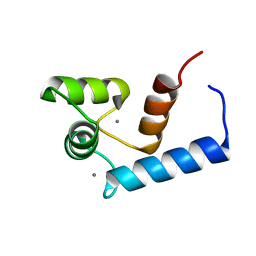 | | Solution structure of calcium bound soybean calmodulin isoform 1 N-terminal domain | | 分子名称: | CALCIUM ION, Calmodulin | | 著者 | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | 登録日 | 2008-03-14 | | 公開日 | 2008-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
8BSU
 
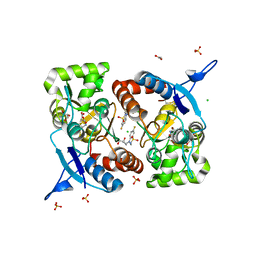 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate and the positive allosteric modulator BPAM344 at 2.9A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2022-11-26 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
8BST
 
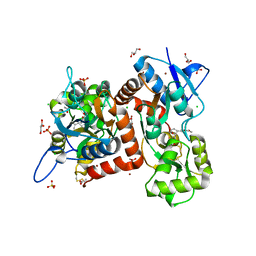 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate at 2.7A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2022-11-26 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
1YGE
 
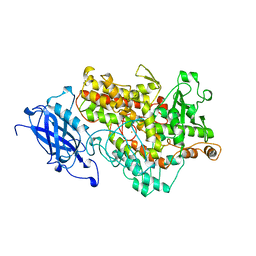 | | LIPOXYGENASE-1 (SOYBEAN) AT 100K | | 分子名称: | FE (III) ION, LIPOXYGENASE-1 | | 著者 | Minor, W, Steczko, J, Stec, B, Otwinowski, Z, Bolin, J.T, Walter, R, Axelrod, B. | | 登録日 | 1996-06-04 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution.
Biochemistry, 35, 1996
|
|
3GBB
 
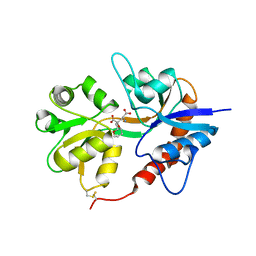 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with MSVIII-19 at 2.10A resolution | | 分子名称: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, Glutamate receptor, ionotropic kainate 1 | | 著者 | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-02-19 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
2BBI
 
 | |
2XI6
 
 | | The structure of ascorbate peroxidase Compound I | | 分子名称: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Gumiero, A, Raven, E.L, Moody, P.C.E. | | 登録日 | 2010-06-29 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
7S10
 
 | | Crystal Structure of ascorbate peroxidase triple mutant: S160M, L203M, Q204M | | 分子名称: | L-ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Poulos, T.L, Kim, J, Murarka, V.C. | | 登録日 | 2021-08-31 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (1.40000689 Å) | | 主引用文献 | Computational analysis of the tryptophan cation radical energetics in peroxidase Compound I.
J.Biol.Inorg.Chem., 27, 2022
|
|
7SOJ
 
 | |
7SOI
 
 | |
7BI1
 
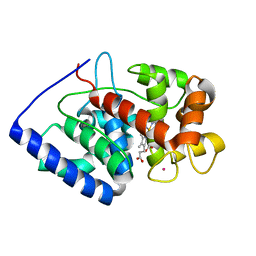 | | XFEL crystal structure of soybean ascorbate peroxidase compound II | | 分子名称: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kwon, H, Tosha, T, Sugimoto, H, Raven, E.L, Moody, P.C.E. | | 登録日 | 2021-01-12 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | XFEL Crystal Structures of Peroxidase Compound II.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2FTL
 
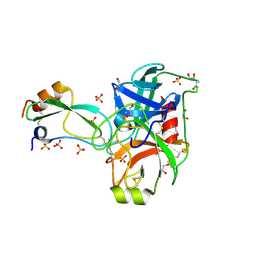 | | Crystal structure of trypsin complexed with BPTI at 100K | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | 著者 | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | 登録日 | 2006-01-24 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
8TQF
 
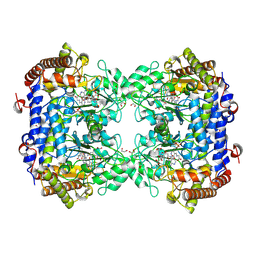 | | Crystal structure of Soybean SHMT8 in complex with PLP-glycine and diglutamylated 5-formyltetrahydrofolate | | 分子名称: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6R)-2-amino-5-formyl-4-hydroxy-5,6,7,8-tetrahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-glutamic acid, ... | | 著者 | Owuocha, L.F, Beamer, L.J. | | 登録日 | 2023-08-07 | | 公開日 | 2024-08-21 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural insights into binding of polyglutamylated tetrahydrofolate by serine hydroxymethyltransferase 8 from soybean.
Front Plant Sci, 15, 2024
|
|
3ZSH
 
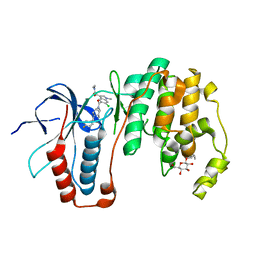 | | X-ray structure of p38alpha bound to SCIO-469 | | 分子名称: | 2-(6-chloro-5-{[(2R,5S)-4-(4-fluorobenzyl)-2,5-dimethylpiperazin-1-yl]carbonyl}-1-methyl-1H-indol-3-yl)-N,N-dimethyl-2-oxoacetamide, MITOGEN-ACTIVATED PROTEIN KINASE 14, octyl beta-D-glucopyranoside | | 著者 | Azevedo, R, van Zeeland, M, Raaijmakers, H, Kazemier, B, Oubrie, A. | | 登録日 | 2011-06-28 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3ZSG
 
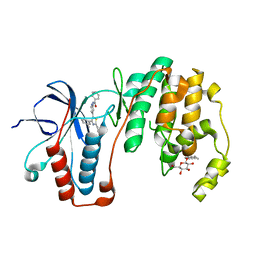 | | X-ray structure of p38alpha bound to TAK-715 | | 分子名称: | MITOGEN-ACTIVATED PROTEIN KINASE 14, TAK-715, octyl beta-D-glucopyranoside | | 著者 | Azevedo, R, van Zeeland, M, Raaijmakers, H, Kazemier, B, Oubrie, A. | | 登録日 | 2011-06-28 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3H6T
 
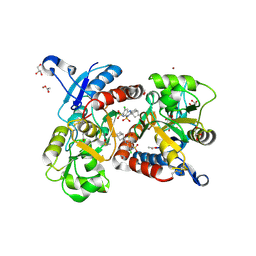 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution | | 分子名称: | ACETATE ION, CACODYLATE ION, CYCLOTHIAZIDE, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6V
 
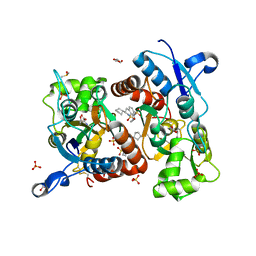 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | 分子名称: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
2VDH
 
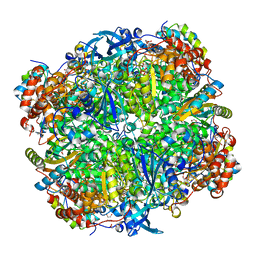 | | Crystal structure of Chlamydomonas reinhardtii Rubisco with a large- subunit C172S mutation | | 分子名称: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Garcia-Murria, M.-J, Karkehabadi, S, Marin-Navarro, J, Satagopan, S, Andersson, I, Spreitzer, R.J, Moreno, J. | | 登録日 | 2007-10-09 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Functional Consequences of the Replacement of Proximal Residues Cys-172 and Cys-192 in the Large Subunit of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase from Chlamydomonas Reinhardtii
Biochem.J., 411, 2008
|
|
7QGF
 
 | | Cubic Insulin SAD phasing at 14.2 keV | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Nanao, M.H, Basu, S. | | 登録日 | 2021-12-08 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.203 Å) | | 主引用文献 | ID23-2: an automated and high-performance microfocus beamline for macromolecular crystallography at the ESRF.
J.Synchrotron Radiat., 29, 2022
|
|
3F3R
 
 | | Crystal structure of yeast Thioredoxin1-glutathione mixed disulfide complex | | 分子名称: | GLUTATHIONE, SULFATE ION, Thioredoxin-1 | | 著者 | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | 登録日 | 2008-10-31 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3F3Q
 
 | | Crystal structure of the oxidised form of thioredoxin 1 from saccharomyces cerevisiae | | 分子名称: | Thioredoxin-1, ZINC ION | | 著者 | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | 登録日 | 2008-10-31 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
2FTM
 
 | | Crystal structure of trypsin complexed with the BPTI variant (Tyr35->Gly) | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | 著者 | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | 登録日 | 2006-01-24 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
