3E0C
 
 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | 分子名称: | DNA damage-binding protein 1 | | 著者 | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2008-07-31 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
2CRX
 
 | |
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Varela, P.F, Charbonnier, J.B. | | 登録日 | 2022-03-14 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | 分子名称: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
3W03
 
 | | XLF-XRCC4 complex | | 分子名称: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | 著者 | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2012-10-17 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (8.492 Å) | | 主引用文献 | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
2R9A
 
 | |
3RWR
 
 | |
6PAI
 
 | |
6CIL
 
 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | 分子名称: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | 著者 | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | 登録日 | 2018-02-24 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.15 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
7ODX
 
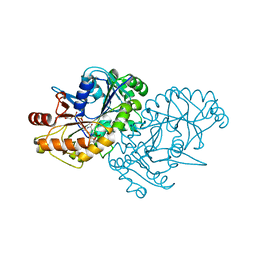 | |
5D2M
 
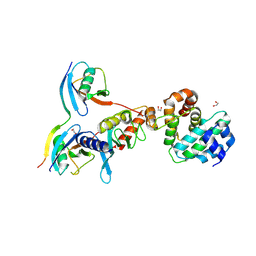 | | Complex between human SUMO2-RANGAP1, UBC9 and ZNF451 | | 分子名称: | 1,2-ETHANEDIOL, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | 著者 | Cappadocia, L, Lima, C.D. | | 登録日 | 2015-08-05 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7PMK
 
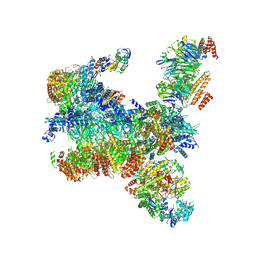 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation I) | | 分子名称: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA helicase, ... | | 著者 | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | 登録日 | 2021-09-02 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7PMN
 
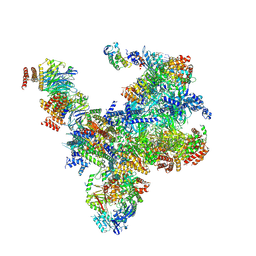 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation II) | | 分子名称: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha-binding protein, ... | | 著者 | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | 登録日 | 2021-09-02 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7BY1
 
 | |
8A58
 
 | |
7V7C
 
 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | 分子名称: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | 著者 | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | 登録日 | 2021-08-21 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
5KGF
 
 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | 分子名称: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | 著者 | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | 登録日 | 2016-06-13 | | 公開日 | 2016-07-27 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.54 Å) | | 主引用文献 | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
6DSZ
 
 | |
6TTU
 
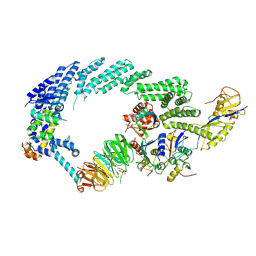 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | 分子名称: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-04 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
4WQO
 
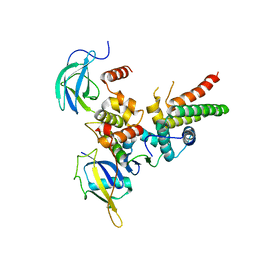 | | Structure of VHL-EloB-EloC-Cul2 | | 分子名称: | Cullin-2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | 著者 | Nguyen, H.C, Xiong, Y. | | 登録日 | 2014-10-22 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Insights into Cullin-RING E3 Ubiquitin Ligase Recruitment: Structure of the VHL-EloBC-Cul2 Complex.
Structure, 23, 2015
|
|
7V7B
 
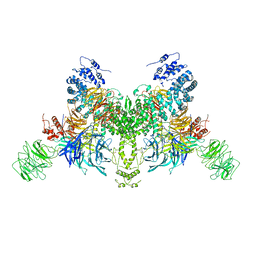 | |
8WQD
 
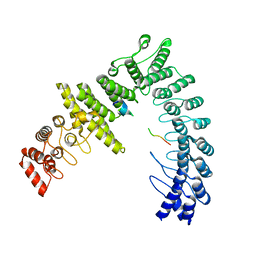 | |
8WQI
 
 | |
