6QUH
 
 | | GHK tagged GFP variant crystal form II at 1.34A wavelength | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, COPPER (II) ION, ... | | 著者 | Huyton, T, Gorlich, D. | | 登録日 | 2019-02-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2022-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2XB6
 
 | | Revisited crystal structure of Neurexin1beta-Neuroligin4 complex | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Leone, P, Comoletti, D, Ferracci, G, Conrod, S, Garcia, S.U, Taylor, P, Bourne, Y, Marchot, P. | | 登録日 | 2010-04-07 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Insights Into the Exquisite Selectivity of Neurexin-Neuroligin Synaptic Interactions
Embo J., 29, 2010
|
|
1HGG
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | 著者 | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | 登録日 | 1991-11-01 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
5FXM
 
 | |
1QKC
 
 | | ESCHERICHIA COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX DELTA TWO-ALBOMYCIN | | 分子名称: | 3-HYDROXY-TETRADECANOIC ACID, DELTA-2-ALBOMYCIN A1, DIPHOSPHATE, ... | | 著者 | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | 登録日 | 1999-07-18 | | 公開日 | 2000-06-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | 分子名称: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | 著者 | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | 登録日 | 2015-11-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
7LQA
 
 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 2 (merged) | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LPU
 
 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 1 | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LN7
 
 | | X-ray radiation damage series on Proteinase K at 277K, crystal structure, dataset 1 | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-06 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6AYF
 
 | | TRPML3/ML-SA1 complex at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-3 | | 著者 | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | 登録日 | 2017-09-08 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7LOQ
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 2 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-10 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LN9
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 1 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-06 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LPV
 
 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 2 | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VFO
 
 | | Crystal structure of SdgB (Phosphate-binding form) | | 分子名称: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LPL
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 3 (merged) | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LQ9
 
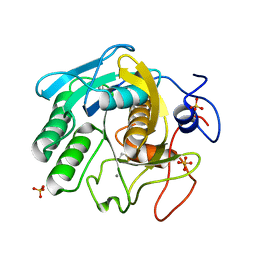 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 4 | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VFM
 
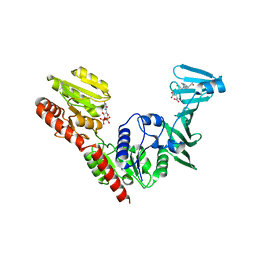 | | Crystal structure of SdgB (UDP and SD peptide-binding form) | | 分子名称: | Glycosyl transferase, group 1 family protein, SER-ASP-SER-ASP, ... | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFN
 
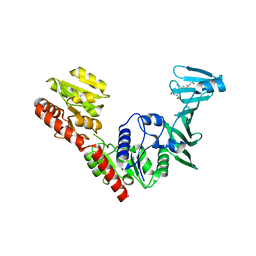 | | Crystal structure of SdgB (SD peptide-binding form) | | 分子名称: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | 著者 | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | 登録日 | 2021-09-13 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6QUJ
 
 | | GHK tagged GFP variant | | 分子名称: | COPPER (II) ION, GLYCEROL, Green fluorescent protein, ... | | 著者 | Huyton, T, Gorlich, D. | | 登録日 | 2019-02-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2022-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7RII
 
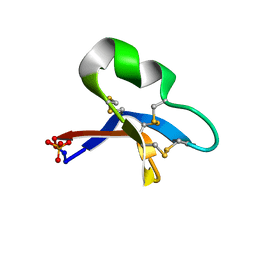 | | [I11L]hyen D crystal structure | | 分子名称: | Cyclotide hyen-D, PHOSPHATE ION | | 著者 | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Enabling efficient folding and high-resolution crystallographic analysis of bracelet cyclotides
Molecules, 26(18), 2021
|
|
7LQB
 
 | | X-ray radiation damage series on Proteinase K at 277K, multi-conformer model, dataset 3 (merged) | | 分子名称: | CALCIUM ION, Proteinase K, SULFATE ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LPM
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, crystal 2 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LOR
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 3 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-10 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LLP
 
 | | X-ray radiation damage series on Lysozyme at 277K, crystal structure, dataset 1 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-04 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LN8
 
 | | X-ray radiation damage series on Lysozyme at 277K, crystal structure, dataset 3 | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yabukarski, F, Doukov, T, Herschlag, D. | | 登録日 | 2021-02-06 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
